+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Thermostabilized human prestin in complex with salicylate | |||||||||
 Map data Map data | postprocess_masked.mrc from RELION | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | motor protein / prestin / SLC26A5 / electromotility / MEMBRANE PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
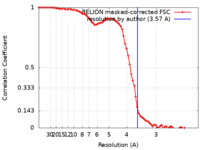
| Method | single particle reconstruction / cryo EM / Resolution: 3.57 Å | |||||||||
 Authors Authors | Futamata H / Fukuda M / Yamashita K / Nishizawa T | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility. Authors: Haon Futamata / Masahiro Fukuda / Rie Umeda / Keitaro Yamashita / Atsuhiro Tomita / Satoe Takahashi / Takafumi Shikakura / Shigehiko Hayashi / Tsukasa Kusakizako / Tomohiro Nishizawa / ...Authors: Haon Futamata / Masahiro Fukuda / Rie Umeda / Keitaro Yamashita / Atsuhiro Tomita / Satoe Takahashi / Takafumi Shikakura / Shigehiko Hayashi / Tsukasa Kusakizako / Tomohiro Nishizawa / Kazuaki Homma / Osamu Nureki /    Abstract: Outer hair cell elecromotility, driven by prestin, is essential for mammalian cochlear amplification. Here, we report the cryo-EM structures of thermostabilized prestin (Pres), complexed with ...Outer hair cell elecromotility, driven by prestin, is essential for mammalian cochlear amplification. Here, we report the cryo-EM structures of thermostabilized prestin (Pres), complexed with chloride, sulfate, or salicylate at 3.52-3.63 Å resolutions. The central positively-charged cavity allows flexible binding of various anion species, which likely accounts for the known distinct modulations of nonlinear capacitance (NLC) by different anions. Comparisons of these Pres structures with recent prestin structures suggest rigid-body movement between the core and gate domains, and provide mechanistic insights into prestin inhibition by salicylate. Mutations at the dimeric interface severely diminished NLC, suggesting that stabilization of the gate domain facilitates core domain movement, thereby contributing to the expression of NLC. These findings advance our understanding of the molecular mechanism underlying mammalian cochlear amplification. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31759.map.gz emd_31759.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31759-v30.xml emd-31759-v30.xml emd-31759.xml emd-31759.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31759_fsc.xml emd_31759_fsc.xml | 7.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_31759.png emd_31759.png | 189.5 KB | ||
| Masks |  emd_31759_msk_1.map emd_31759_msk_1.map | 18.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31759.cif.gz emd-31759.cif.gz | 6.4 KB | ||
| Others |  emd_31759_half_map_1.map.gz emd_31759_half_map_1.map.gz emd_31759_half_map_2.map.gz emd_31759_half_map_2.map.gz | 15.5 MB 15.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31759 http://ftp.pdbj.org/pub/emdb/structures/EMD-31759 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31759 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31759 | HTTPS FTP |
-Validation report
| Summary document |  emd_31759_validation.pdf.gz emd_31759_validation.pdf.gz | 741.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31759_full_validation.pdf.gz emd_31759_full_validation.pdf.gz | 740.9 KB | Display | |
| Data in XML |  emd_31759_validation.xml.gz emd_31759_validation.xml.gz | 13.5 KB | Display | |
| Data in CIF |  emd_31759_validation.cif.gz emd_31759_validation.cif.gz | 17.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31759 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31759 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31759 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31759 | HTTPS FTP |
-Related structure data
| Related structure data |  7v75MC  7v73C  7v74C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31759.map.gz / Format: CCP4 / Size: 18.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31759.map.gz / Format: CCP4 / Size: 18.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess_masked.mrc from RELION | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.10667 Å | ||||||||||||||||||||||||||||||||||||
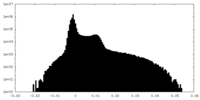
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31759_msk_1.map emd_31759_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
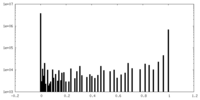
| Density Histograms |
-Half map: run half1 class001 unfil.mrc from RELION
| File | emd_31759_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | run_half1_class001_unfil.mrc from RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
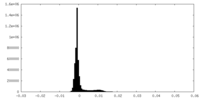
| Density Histograms |
-Half map: run half2 class001 unfil.mrc from RELION
| File | emd_31759_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | run_half2_class001_unfil.mrc from RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
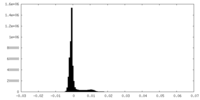
| Density Histograms |
- Sample components
Sample components
-Entire : prestin in complex with chloride ion
| Entire | Name: prestin in complex with chloride ion |
|---|---|
| Components |
|
-Supramolecule #1: prestin in complex with chloride ion
| Supramolecule | Name: prestin in complex with chloride ion / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: prestin
| Macromolecule | Name: prestin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.503375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDHAEENEIL AATQRYVVER PVYSQELLEE ELEKKDRVPK TLGDKLKKSF RCSPKKAKNL LLSFFPILEW LPKYNLKEWL LGDLIAGLT VGSLQIPQGI AFALLAGLPP IYGLYSSFFP PLIYAFFGTS RHISVGPFAV VSLLVGSVVE RLVPDDIVLP G GVNATNGT ...String: MDHAEENEIL AATQRYVVER PVYSQELLEE ELEKKDRVPK TLGDKLKKSF RCSPKKAKNL LLSFFPILEW LPKYNLKEWL LGDLIAGLT VGSLQIPQGI AFALLAGLPP IYGLYSSFFP PLIYAFFGTS RHISVGPFAV VSLLVGSVVE RLVPDDIVLP G GVNATNGT EARDALRVQV AFTLTFLAGI IQLALGLLRL GFLVDFLSEP LISGFTTGAA IHILLSQLKY LLGLKIPRHS GP FSVIYSV ISVFHNIPNT NIATLGVSLL SFVLLLVVKE LNKRFKKKLP VPIPAELIVV ILATLISYYF NLAEKYGVSI VGH IPKGLP PPSVPDLSLF PLVIGDAIAI AIVGLAVSIS VGKTFAKKHG YQIDGNQELI ALGLMNIVGS FFSCYPATGS FSRS AVNES AGGKTQVAGI VAALVVLLVL LFLGPLFYYL PKAVLAAIII VNLKGLLMQF KDAPKLWKVD KLDFLIWLVT FLGVV FLSV EIGLLVGVGF SLLTVLLRTQ RPRTSVLGRI PGTDIYRDVK QYPEAEEVPG VKIFRIDSPI YFANSEYFKE RLKRKT GVD PVKVLAARKK ALKKIEKEIK KANLANKTVV KADAEVDGED ATKPEEEDGE VKYPPIVIQS DWPSELPRFV PPKVDFH TL ILDFSAVSFV DTVGVKTLKE IVKEYREIGV QVYLAGCNAS VVEKLERGGF FDDGITKEHL FLSVHDAVLF AQARKALA E QEASAPPSQE DLEPNATPAT PEAGTENLYF QG |
-Macromolecule #2: 2-HYDROXYBENZOIC ACID
| Macromolecule | Name: 2-HYDROXYBENZOIC ACID / type: ligand / ID: 2 / Number of copies: 1 / Formula: SAL |
|---|---|
| Molecular weight | Theoretical: 138.121 Da |
| Chemical component information | 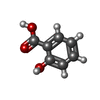 ChemComp-SAL: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #4: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 4 / Number of copies: 8 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)