+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative-staining map of T4 DNA ligase-AMP Binary Complex II | |||||||||
 Map data Map data | T4 DNA ligase conformation Open II | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ligase / DNA Repair / DNA replication / DNA BINDING PROTEIN | |||||||||
| Biological species |  bacteriophage T (virus) bacteriophage T (virus) | |||||||||
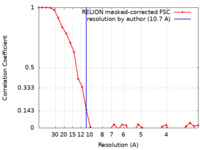
| Method | single particle reconstruction / negative staining / Resolution: 10.7 Å | |||||||||
 Authors Authors | Zhang L / Li N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Repair of DNA nick by T4 DNA ligase in three parallel pathways Authors: Zhang L / Li N | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31751.map.gz emd_31751.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31751-v30.xml emd-31751-v30.xml emd-31751.xml emd-31751.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31751_fsc.xml emd_31751_fsc.xml | 3 KB | Display |  FSC data file FSC data file |
| Images |  emd_31751.png emd_31751.png | 31.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31751 http://ftp.pdbj.org/pub/emdb/structures/EMD-31751 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31751 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31751 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31751.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31751.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T4 DNA ligase conformation Open II | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.607 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : T4 DNA ligase conformation Open II
| Entire | Name: T4 DNA ligase conformation Open II |
|---|---|
| Components |
|
-Supramolecule #1: T4 DNA ligase conformation Open II
| Supramolecule | Name: T4 DNA ligase conformation Open II / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  bacteriophage T (virus) bacteriophage T (virus) |
| Molecular weight | Theoretical: 55 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.017 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: A 4.0 uL drop of the sample was placed on a glow-discharged EM specimen grid (CF200-Cu, Electron Microscopy Sciences, Hatfield, PA, USA) with a thin carbon film for 1 min and the excess ...Details: A 4.0 uL drop of the sample was placed on a glow-discharged EM specimen grid (CF200-Cu, Electron Microscopy Sciences, Hatfield, PA, USA) with a thin carbon film for 1 min and the excess solution was blotted with filter paper. Then, the grid was washed with a clean drop (~35 uL/drop) of water and stained two times with two drops (~25 uL/drop) of 2% (w/v) uranyl acetate (UA) for 10 s, 30 s respectively before air-drying. | ||||||||||||||||||
| Grid | Model: C-flat / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.038 kPa |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS F200C |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 67 / Average exposure time: 1.0 sec. / Average electron dose: 160.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)