[English] 日本語
 Yorodumi
Yorodumi- EMDB-31711: Cryo-EM Structure of Glycine max glutamine synthetase GmGS Beta2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Glycine max glutamine synthetase GmGS Beta2 | |||||||||
 Map data Map data | GmGS Beta2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | supramolecular enzyme / glutamine synthetase / Glycine max / PLANT PROTEIN / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamine synthetase / glutamine biosynthetic process / glutamine synthetase activity / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Xu W / Chen Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Assembly status transition offers an avenue for activity modulation of a supramolecular enzyme. Authors: Yao Chen / Weiya Xu / Shuwei Yu / Kang Ni / Guangbiao She / Xiaodong Ye / Qiong Xing / Jian Zhao / Chengdong Huang /  Abstract: Nature has evolved many supramolecular proteins assembled in certain, sometimes even seemingly oversophisticated, morphological manners. The rationale behind such evolutionary efforts is often poorly ...Nature has evolved many supramolecular proteins assembled in certain, sometimes even seemingly oversophisticated, morphological manners. The rationale behind such evolutionary efforts is often poorly understood. Here, we provide atomic-resolution insights into how the dynamic building of a structurally complex enzyme with higher order symmetry offers amenability to intricate regulation. We have established the functional coupling between enzymatic activity and protein morphological states of glutamine synthetase (GS), an old multi-subunit enzyme essential for cellular nitrogen metabolism. Cryo-EM structure determination of GS in both the catalytically active and inactive assembly states allows us to reveal an unanticipated self-assembly-induced disorder-order transition paradigm, in which the remote interactions between two subcomplex entities significantly rigidify the otherwise structurally fluctuating active sites, thereby regulating activity. We further show in vivo evidences that how the enzyme morphology transitions could be modulated by cellular factors on demand. Collectively, our data present an example of how assembly status transition offers an avenue for activity modulation, and sharpens our mechanistic understanding of the complex functional and regulatory properties of supramolecular enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31711.map.gz emd_31711.map.gz | 482.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31711-v30.xml emd-31711-v30.xml emd-31711.xml emd-31711.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31711.png emd_31711.png | 239.7 KB | ||
| Filedesc metadata |  emd-31711.cif.gz emd-31711.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31711 http://ftp.pdbj.org/pub/emdb/structures/EMD-31711 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31711 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31711 | HTTPS FTP |
-Validation report
| Summary document |  emd_31711_validation.pdf.gz emd_31711_validation.pdf.gz | 562.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31711_full_validation.pdf.gz emd_31711_full_validation.pdf.gz | 561.8 KB | Display | |
| Data in XML |  emd_31711_validation.xml.gz emd_31711_validation.xml.gz | 8 KB | Display | |
| Data in CIF |  emd_31711_validation.cif.gz emd_31711_validation.cif.gz | 9.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31711 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31711 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31711 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31711 | HTTPS FTP |
-Related structure data
| Related structure data |  7v4hMC  7v4iC  7v4jC  7v4kC  7v4lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31711.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31711.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GmGS Beta2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.505 Å | ||||||||||||||||||||||||||||||||||||
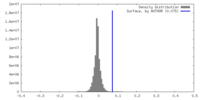
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : GmGS Beta2
| Entire | Name: GmGS Beta2 |
|---|---|
| Components |
|
-Supramolecule #1: GmGS Beta2
| Supramolecule | Name: GmGS Beta2 / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamine synthetase
| Macromolecule | Name: Glutamine synthetase / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO / EC number: glutamine synthetase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.184859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLLSDLINL NLSDTTEKVI AEYIWIGGSG MDLRSKARTL PGPVSDPSKL PKWNYDGSST GQAPGEDSEV IIYPQAIFRD PFRRGNNIL VICDTYTPAG EPIPTNKRHD AAKVFSHPDV VAEETWYGIE QEYTLLQKDI QWPLGWPVGG FPGPQGPYYC G VGADKAFG ...String: MSLLSDLINL NLSDTTEKVI AEYIWIGGSG MDLRSKARTL PGPVSDPSKL PKWNYDGSST GQAPGEDSEV IIYPQAIFRD PFRRGNNIL VICDTYTPAG EPIPTNKRHD AAKVFSHPDV VAEETWYGIE QEYTLLQKDI QWPLGWPVGG FPGPQGPYYC G VGADKAFG RDIVDAHYKA CLYAGINISG INGEVMPGQW EFQVGPSVGI SAGDEVWAAR YILERITEIA GVVVSFDPKP IQ GDWNGAG AHTNYSTKSM RNDGGYEVIK TAIEKLGKRH KEHIAAYGEG NERRLTGRHE TADINTFLWG VANRGASVRV GRD TEKAGK GYFEDRRPAS NMDPYVVTSM IADTTILWKP UniProtKB: Glutamine synthetase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


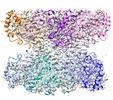





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)