+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM structure of lysosomal ATPase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | lysosomal ATPase transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpolyamine transmembrane transporter activity / polyamine transmembrane transport / spermine transmembrane transport / : / ABC-type polyamine transporter activity / regulation of autophagosome size / extracellular exosome biogenesis / negative regulation of lysosomal protein catabolic process / regulation of chaperone-mediated autophagy / P-type ion transporter activity ...polyamine transmembrane transporter activity / polyamine transmembrane transport / spermine transmembrane transport / : / ABC-type polyamine transporter activity / regulation of autophagosome size / extracellular exosome biogenesis / negative regulation of lysosomal protein catabolic process / regulation of chaperone-mediated autophagy / P-type ion transporter activity / regulation of autophagy of mitochondrion / regulation of lysosomal protein catabolic process / intracellular monoatomic cation homeostasis / autophagosome-lysosome fusion / autophagosome organization / protein localization to lysosome / positive regulation of exosomal secretion / phosphatidic acid binding / ATPase-coupled monoatomic cation transmembrane transporter activity / multivesicular body membrane / intracellular zinc ion homeostasis / cupric ion binding / regulation of protein localization to nucleus / regulation of mitochondrion organization / phosphatidylinositol-3,5-bisphosphate binding / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / lysosomal transport / regulation of intracellular protein transport / cellular response to zinc ion / lipid homeostasis / autophagosome membrane / Ion transport by P-type ATPases / regulation of macroautophagy / transport vesicle / regulation of neuron apoptotic process / cellular response to manganese ion / multivesicular body / lysosomal lumen / autophagosome / positive regulation of protein secretion / autophagy / intracellular calcium ion homeostasis / late endosome membrane / late endosome / manganese ion binding / cellular response to oxidative stress / monoatomic ion transmembrane transport / vesicle / intracellular iron ion homeostasis / lysosome / neuron projection / lysosomal membrane / neuronal cell body / positive regulation of gene expression / ATP hydrolysis activity / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhang SS / Yang MJ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2021 Journal: Cell Discov / Year: 2021Title: Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2. Authors: Xudong Chen / Mingze Zhou / Sensen Zhang / Jian Yin / Ping Zhang / Xujun Xuan / Peiyi Wang / Zhiqiang Liu / Boda Zhou / Maojun Yang /  Abstract: Polyamines are important polycations that play critical roles in mammalian cells. ATP13A2 belongs to the orphan P5B adenosine triphosphatases (ATPase) family and has been established as a lysosomal ...Polyamines are important polycations that play critical roles in mammalian cells. ATP13A2 belongs to the orphan P5B adenosine triphosphatases (ATPase) family and has been established as a lysosomal polyamine exporter to maintain the normal function of lysosomes and mitochondria. Previous studies have reported that several human neurodegenerative disorders are related to mutations in the ATP13A2 gene. However, the transport mechanism of ATP13A2 in the lysosome remains unclear. Here, we report the cryo-electron microscopy (cryo-EM) structures of three distinct intermediates of the human ATP13A2, revealing key insights into the spermine (SPM) transport cycle in the lysosome. The transmembrane domain serves as a substrate binding site and the C-terminal domain is essential for protein stability and may play a regulatory role. These findings advance our understanding of the polyamine transport mechanism, the lipid-associated regulation, and the disease-associated mutants of ATP13A2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31623.map.gz emd_31623.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31623-v30.xml emd-31623-v30.xml emd-31623.xml emd-31623.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31623.png emd_31623.png | 45.1 KB | ||
| Filedesc metadata |  emd-31623.cif.gz emd-31623.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31623 http://ftp.pdbj.org/pub/emdb/structures/EMD-31623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31623 | HTTPS FTP |
-Related structure data
| Related structure data |  7fjmMC  7fjpC  7fjqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31623.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31623.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0979 Å | ||||||||||||||||||||||||||||||||||||
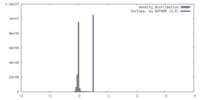
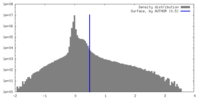
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : plasma membaren channel
| Entire | Name: plasma membaren channel |
|---|---|
| Components |
|
-Supramolecule #1: plasma membaren channel
| Supramolecule | Name: plasma membaren channel / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Polyamine-transporting ATPase 13A2
| Macromolecule | Name: Polyamine-transporting ATPase 13A2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 125.367961 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSADSSPLVG STPTGYGTLT IGTSIDPLSS SVSSVRLSGY CGSPWRVIGY HVVVWMMAGI PLLLFRWKPL WGVRLRLRPC NLAHAETLV IEIRDKEDSS WQLFTVQVQT EAIGEGSLEP SPQSQAEDGR SQAAVGAVPE GAWKDTAQLH KSEEAVSVGQ K RVLRYYLF ...String: MSADSSPLVG STPTGYGTLT IGTSIDPLSS SVSSVRLSGY CGSPWRVIGY HVVVWMMAGI PLLLFRWKPL WGVRLRLRPC NLAHAETLV IEIRDKEDSS WQLFTVQVQT EAIGEGSLEP SPQSQAEDGR SQAAVGAVPE GAWKDTAQLH KSEEAVSVGQ K RVLRYYLF QGQRYIWIET QQAFYQVSLL DHGRSCDDVH RSRHGLSLQD QMVRKAIYGP NVISIPVKSY PQLLVDEALN PY YGFQAFS IALWLADHYY WYALCIFLIS SISICLSLYK TRKQSQTLRD MVKLSMRVCV CRPGGEEEWV DSSELVPGDC LVL PQEGGL MPCDAALVAG ECMVNESSLT GESIPVLKTA LPEGLGPYCA ETHRRHTLFC GTLILQARAY VGPHVLAVVT RTGF CTAKG GLVSSILHPR PINFKFYKHS MKFVAALSVL ALLGTIYSIF ILYRNRVPLN EIVIRALDLV TVVVPPALPA AMTVC TLYA QSRLRRQGIF CIHPLRINLG GKLQLVCFDK TGTLTEDGLD VMGVVPLKGQ AFLPLVPEPR RLPVGPLLRA LATCHA LSR LQDTPVGDPM DLKMVESTGP QLQAMEEPPV PVSVLHRFPF SSALQRMSVV VAWPGATQPE AYVKGSPELV AGLCNPE TV PTDFAQMLQS YTAAGYRVVA LASKPLPTVP SLEAAQQLTR DTVEGDLSLL GLLVMRNLLK PQTTPVIQAL RRTRIRAV M VTGDNLQTAV TVARGCGMVA PQEHLIIVHA TGQPASLEFL PMESPTAVNG VKDPDQAASY TVEPDPRSRH LALSGPTFG IIVKHFPKLL PKVLVQGTVF ARMAPEQKTE LVCELQKLQY CVGMCGDGAN DCGALKAADV GISLSQAEAS VVSPFTSSMA SIECVPMVI REGRCSLDTS FSVFKYMALY SLTQFISVLI LYTINTNLGD LQFLAIDLVI TTTVAVLMSR TGPALVLGRV R PPGALLSV PVLSSLLLQM VLVTGVQLGG YFLTLAQPWF VPLNRTVAAP DNLPNYENTV VFSLSSFQYL ILAAAVSKGA PF RRPLYTN VPFLVALALL SSVLVGLVLV PGLLQGPLAL RNITDTGFKL LLLGLVTLNF VGAFMLESVL DQCLPACLRR LRP KRASKK RFKQLERELA EQPWPPLPAG PLR UniProtKB: Polyamine-transporting ATPase 13A2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 153193 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)