+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human NTCP in complex with YN69083 Fab | |||||||||
 Map data Map data | postprocess map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter / TRANSPORT PROTEIN / TRANSPORT PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationbile acid:sodium symporter activity / bile acid transmembrane transporter activity / bile acid and bile salt transport / Recycling of bile acids and salts / response to nutrient levels / response to estrogen / cellular response to xenobiotic stimulus / virus receptor activity / response to ethanol / basolateral plasma membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
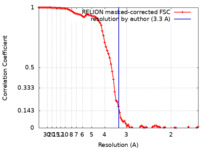
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Park JH / Iwamoto M | |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structural insights into the HBV receptor and bile acid transporter NTCP. Authors: Jae-Hyun Park / Masashi Iwamoto / Ji-Hye Yun / Tomomi Uchikubo-Kamo / Donghwan Son / Zeyu Jin / Hisashi Yoshida / Mio Ohki / Naito Ishimoto / Kenji Mizutani / Mizuki Oshima / Masamichi ...Authors: Jae-Hyun Park / Masashi Iwamoto / Ji-Hye Yun / Tomomi Uchikubo-Kamo / Donghwan Son / Zeyu Jin / Hisashi Yoshida / Mio Ohki / Naito Ishimoto / Kenji Mizutani / Mizuki Oshima / Masamichi Muramatsu / Takaji Wakita / Mikako Shirouzu / Kehong Liu / Tomoko Uemura / Norimichi Nomura / So Iwata / Koichi Watashi / Jeremy R H Tame / Tomohiro Nishizawa / Weontae Lee / Sam-Yong Park /   Abstract: Around 250 million people are infected with hepatitis B virus (HBV) worldwide, and 15 million may also carry the satellite virus hepatitis D virus (HDV), which confers even greater risk of severe ...Around 250 million people are infected with hepatitis B virus (HBV) worldwide, and 15 million may also carry the satellite virus hepatitis D virus (HDV), which confers even greater risk of severe liver disease. The HBV receptor has been identified as sodium taurocholate co-transporting polypeptide (NTCP), which interacts directly with the first 48 amino acid residues of the N-myristoylated N-terminal preS1 domain of the viral large protein. Despite the pressing need for therapeutic agents to counter HBV, the structure of NTCP remains unsolved. This 349-residue protein is closely related to human apical sodium-dependent bile acid transporter (ASBT), another member of the solute carrier family SLC10. Crystal structures have been reported of similar bile acid transporters from bacteria, and these models are believed to resemble closely both NTCP and ASBT. Here we have used cryo-electron microscopy to solve the structure of NTCP bound to an antibody, clearly showing that the transporter has no equivalent of the first transmembrane helix found in other SLC10 proteins, and that the N terminus is exposed on the extracellular face. Comparison of our structure with those of related proteins indicates a common mechanism of bile acid transport, but the NTCP structure displays an additional pocket formed by residues that are known to interact with preS1, presenting new opportunities for structure-based drug design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31526.map.gz emd_31526.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31526-v30.xml emd-31526-v30.xml emd-31526.xml emd-31526.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31526_fsc.xml emd_31526_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_31526.png emd_31526.png | 102.4 KB | ||
| Masks |  emd_31526_msk_1.map emd_31526_msk_1.map | 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31526.cif.gz emd-31526.cif.gz | 6.3 KB | ||
| Others |  emd_31526_half_map_1.map.gz emd_31526_half_map_1.map.gz emd_31526_half_map_2.map.gz emd_31526_half_map_2.map.gz | 52 MB 52 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31526 http://ftp.pdbj.org/pub/emdb/structures/EMD-31526 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31526 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31526 | HTTPS FTP |
-Related structure data
| Related structure data |  7fciMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11178 (Title: Human NTCP in complex with YN69083 Fab / Data size: 5.5 TB EMPIAR-11178 (Title: Human NTCP in complex with YN69083 Fab / Data size: 5.5 TBData #1: Unaligned, non-dose weighted movies of NTCP-YN69083Fab Data 1 [micrographs - multiframe] Data #2: Unaligned, non-dose weighted movies of NTCP-YN69083Fab Data 2 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31526.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31526.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.829 Å | ||||||||||||||||||||||||||||||||||||
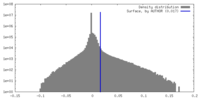
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31526_msk_1.map emd_31526_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
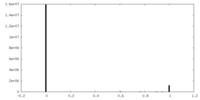
| Density Histograms |
-Half map: halfmap1
| File | emd_31526_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
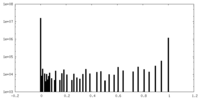
| Density Histograms |
-Half map: halfmap2
| File | emd_31526_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
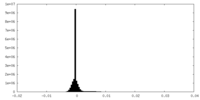
| Density Histograms |
- Sample components
Sample components
-Entire : human NTCP in complex with YN69083
| Entire | Name: human NTCP in complex with YN69083 |
|---|---|
| Components |
|
-Supramolecule #1: human NTCP in complex with YN69083
| Supramolecule | Name: human NTCP in complex with YN69083 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 89 KDa |
-Supramolecule #2: NTCP
| Supramolecule | Name: NTCP / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Fab Light chain,Fab Heavy chain
| Supramolecule | Name: Fab Light chain,Fab Heavy chain / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Sodium/bile acid cotransporter
| Macromolecule | Name: Sodium/bile acid cotransporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.529355 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGAPMEAHN ASAPFNFTLP PNFGKRPTDL ALSVILVFML FFIMLSLGCT MEFSKIKAHL WKPKGLAIAL VAQYGIMPLT AFVLGKVFR LKNIEALAIL VCGCSPGGNL SNVFSLAMKG DMNLSIVMTT CSTFCALGMM PLLLYIYSRG IYDGDLKDKV P YKGIVISL ...String: GPGAPMEAHN ASAPFNFTLP PNFGKRPTDL ALSVILVFML FFIMLSLGCT MEFSKIKAHL WKPKGLAIAL VAQYGIMPLT AFVLGKVFR LKNIEALAIL VCGCSPGGNL SNVFSLAMKG DMNLSIVMTT CSTFCALGMM PLLLYIYSRG IYDGDLKDKV P YKGIVISL VLVLIPCTIG IVLKSKRPQY MRYVIKGGMI IILLCSVAVT VLSAINVGKS IMFAMTPLLI ATSSLMPFIG FL LGYVLSA LFCLNGRCRR TVSMETGCQN VQLCSTILNV AFPPEVIGPL FFFPLLYMIF QLGEGLLLIA IFWCYEKFKT PKD KTKMIY TAATTEETIP GALGNGTYKG EDCSPCTA UniProtKB: Hepatic sodium/bile acid cotransporter |
-Macromolecule #2: Fab Light chain
| Macromolecule | Name: Fab Light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.3829 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVMTQSPAI MSASPGQKVT ITCSASSSVN YMHWYQQKLG SSPKLWIYDT SKLALGVPAR FSGSGSGTSY SLTISSMEAE DAASYFCHQ WSSYPRTFGG GTKLEIKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK ...String: DIVMTQSPAI MSASPGQKVT ITCSASSSVN YMHWYQQKLG SSPKLWIYDT SKLALGVPAR FSGSGSGTSY SLTISSMEAE DAASYFCHQ WSSYPRTFGG GTKLEIKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK DSTYSMSSTL TLTKDEYERH NSYTCEATHK TSTSPIVKSF NRNEC |
-Macromolecule #3: Fab Heavy chain
| Macromolecule | Name: Fab Heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.418123 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLQQPGAE LVKPGASVKL SCKTSGYTFT NYWMKWVKQR PGQGLEWIGE INPSNGGTNY NGKFKSKASL TVDKSSSTAY MQLSSLTSE DSAVYYCTIL VYDAYYVFAM DYWGLGTSVT VSSAKTTPPS VYPLAPGSAA QTNSMVTLGC LVKGYFPEPV T VTWNSGSL ...String: EVQLQQPGAE LVKPGASVKL SCKTSGYTFT NYWMKWVKQR PGQGLEWIGE INPSNGGTNY NGKFKSKASL TVDKSSSTAY MQLSSLTSE DSAVYYCTIL VYDAYYVFAM DYWGLGTSVT VSSAKTTPPS VYPLAPGSAA QTNSMVTLGC LVKGYFPEPV T VTWNSGSL SSGVHTFPAV LQSDLYTLSS SVTVPSSTWP SETVTCNVAH PASSTKVDKK IVPRDCGCKP CICTVPEVSS VF IFPPKPK DVLTITLT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 302 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)