[English] 日本語
 Yorodumi
Yorodumi- EMDB-31509: SARS-CoV-2 spike protein in one-RBD open state after CTSL treatment -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike protein in one-RBD open state after CTSL treatment | |||||||||
 Map data Map data | SARS-CoV-2 spike protein in one-RBD open state after CTSL treatment | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
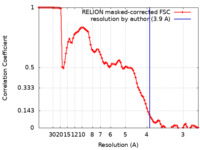
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Zhu Y / Tai L / Sun F | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Novel sites for Cathepsin L cleavage in SARS-CoV-2 spike guide treatment strategies Authors: Zhao M / Zhu Y / Tai L / Yin G / Sun F / Yang J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31509.map.gz emd_31509.map.gz | 58.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31509-v30.xml emd-31509-v30.xml emd-31509.xml emd-31509.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31509_fsc.xml emd_31509_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_31509.png emd_31509.png | 9.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31509 http://ftp.pdbj.org/pub/emdb/structures/EMD-31509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31509 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31509.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31509.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 spike protein in one-RBD open state after CTSL treatment | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Severe acute respiratory syndrome coronavirus 2
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Severe acute respiratory syndrome coronavirus 2
| Supramolecule | Name: Severe acute respiratory syndrome coronavirus 2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2697049 Sci species name: Severe acute respiratory syndrome coronavirus 2 Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host system | Organism:  Homo sapiens (human) / Recombinant cell: Expi293 Homo sapiens (human) / Recombinant cell: Expi293 |
-Macromolecule #1: SARS-CoV-2 spike protein
| Macromolecule | Name: SARS-CoV-2 spike protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MFVFLVLLP LVSSQCVNLT TRTQLPPAYT NSFTRGVYYP DKVFRSSVLH STQDLFLPFF SNVTWFHAIH VSGTNGTKRF DNPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV YSSANNCTFE ...String: MFVFLVLLP LVSSQCVNLT TRTQLPPAYT NSFTRGVYYP DKVFRSSVLH STQDLFLPFF SNVTWFHAIH VSGTNGTKRF DNPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV YSSANNCTFE YVSQPFLMDL EGKQGNFKNL REFVFKNIDG YFKIYSKHTP INLVRDLPQG FSALEPLVDL PIGINITRFQ TLLALHRSYL TPGDSSSGWT AGAAAYYVGY LQPRTFLLKY NENGTITDAV DCALDPLSET KCTLKSFTVE KGIYQTSNFR VQPTESIVRF PNITNLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIRGDEVRQ IAPGQTGKIA DYNYKLPDDF TGCVIAWNSN NLDSKVGGNY NYLYRLFRKS NLKPFERDIS TEIYQAGSTP CNGVEGFNCY FPLQSYGFQP TNGVGYQPYR VVVLSFELLH APATVCGPKK STNLVKNKCV NFNFNGLTGT GVLTESNKKF LPFQQFGRDI ADTTDAVRDP QTLEILDITP CSFGGVSVIT PGTNTSNQVA VLYQDVNCTE VPVAIHADQL TPTWRVYSTG SNVFQTRAGC LIGAEHVNNS YECDIPIGAG ICASYQTQTN SPGAASSVAS QSIIAYTMSL GAENSVAYSN NSIAIPTNFT ISVTTEILPV SMTKTSVDCT MYICGDSTEC SNLLLQYGSF CTQLNRALTG IAVEQDKNTQ EVFAQVKQIY KTPPIKDFGG FNFSQILPDP SKPSKRSFIE DLLFNKVTLA DAGFIKQYGD CLGDIAARDL ICAQKFNGLT VLPPLLTDEM IAQYTSALLA GTITSGWTFG AGAALQIPFA MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STASALGKLQ DVVNQNAQAL NTLVKQLSSN FGAISSVLND ILSRLDPPEA EVQIDRLITG RLQSLQTYVT QQLIRAAEIR ASANLAATKM SECVLGQSKR VDFCGKGYHL MSFPQSAPHG VVFLHVTYVP AQEKNFTTAP AICHDGKAHF PREGVFVSNG THWFVTQRNF YEPQIITTDN TFVSGNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQELGKYEQL VPRGSGYIPE APRDGQAYVR KDGEWVLLST FLHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)