[English] 日本語
 Yorodumi
Yorodumi- EMDB-31446: Cryo-EM map of Rhizobium etli MprF with the improved density of t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of Rhizobium etli MprF with the improved density of the aa-PGS domain | |||||||||
 Map data Map data | 3.3 A Refine3D map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Rhizobium etli CFN 42 (bacteria) Rhizobium etli CFN 42 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Nishimura M / Hirano H / Kobayashi K / Gill CP / Phan CNK / Kise Y / Kusakizako T / Yamashita K / Ito Y / Roy H ...Nishimura M / Hirano H / Kobayashi K / Gill CP / Phan CNK / Kise Y / Kusakizako T / Yamashita K / Ito Y / Roy H / Nishizawa T / Nureki O | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: to be published Authors: Nishimura M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31446.map.gz emd_31446.map.gz | 23.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31446-v30.xml emd-31446-v30.xml emd-31446.xml emd-31446.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31446_fsc.xml emd_31446_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_31446.png emd_31446.png | 144 KB | ||
| Masks |  emd_31446_msk_1.map emd_31446_msk_1.map | 28.7 MB |  Mask map Mask map | |
| Others |  emd_31446_half_map_1.map.gz emd_31446_half_map_1.map.gz emd_31446_half_map_2.map.gz emd_31446_half_map_2.map.gz | 24.1 MB 24.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31446 http://ftp.pdbj.org/pub/emdb/structures/EMD-31446 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31446 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31446 | HTTPS FTP |
-Validation report
| Summary document |  emd_31446_validation.pdf.gz emd_31446_validation.pdf.gz | 685.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31446_full_validation.pdf.gz emd_31446_full_validation.pdf.gz | 685.3 KB | Display | |
| Data in XML |  emd_31446_validation.xml.gz emd_31446_validation.xml.gz | 14.6 KB | Display | |
| Data in CIF |  emd_31446_validation.cif.gz emd_31446_validation.cif.gz | 18.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31446 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31446 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31446 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31446 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31446.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31446.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3.3 A Refine3D map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.996 Å | ||||||||||||||||||||||||||||||||||||
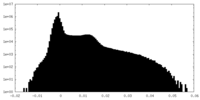
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31446_msk_1.map emd_31446_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
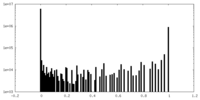
| Density Histograms |
-Half map: #1
| File | emd_31446_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
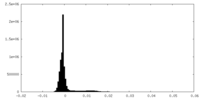
| Density Histograms |
-Half map: #2
| File | emd_31446_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
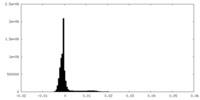
| Density Histograms |
- Sample components
Sample components
-Entire : MprF
| Entire | Name: MprF |
|---|---|
| Components |
|
-Supramolecule #1: MprF
| Supramolecule | Name: MprF / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Rhizobium etli CFN 42 (bacteria) Rhizobium etli CFN 42 (bacteria) |
| Molecular weight | Theoretical: 187 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #1: MprF
| Macromolecule | Name: MprF / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhizobium etli CFN 42 (bacteria) Rhizobium etli CFN 42 (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MSGHGNLEEM EATDGFSFRT LFRRYRTPLT AAATLIVFCL VGYAIMQLTN EVRYDDVVAA LAATGPSAIL LALFFTALSF LSLVFYDLNA IEYIGKKLPF PHVALTAFSA YAVGNTAGFG ALSGGAIRYR AYTRLGLSPE DIGRIIAFVT LSFGLGLAAV ASIALIIIAS ...String: MSGHGNLEEM EATDGFSFRT LFRRYRTPLT AAATLIVFCL VGYAIMQLTN EVRYDDVVAA LAATGPSAIL LALFFTALSF LSLVFYDLNA IEYIGKKLPF PHVALTAFSA YAVGNTAGFG ALSGGAIRYR AYTRLGLSPE DIGRIIAFVT LSFGLGLAAV ASIALIIIAS EIGPLIGVSP FLLRLIAGSI IAILGAVMII GREGRVLNFG AVAIRLPDSR TWSRQFLVTA FDIAASASVL YVLLPQTAIG WPVFLAVYAI AVGLGVLSHV PAGLGVFETV IIASLGSAVN IDAVLGSLVL YRLIYHVLPL LIAVLAVSAA ELRRFVDHPA ASSVRRIGGR LMPQLLSTLA LLLGVMLVFS SVTPTPDQNL EFLSNYLPLP MVEGAHFLSS LLGLALVVAA RGLGQRLDGA WWVAVFSAVA ALTLSLLKAI ALVEAAFLAF LIFGLFVSRR LFTRHASLLN QAMTASWLMA IAVIVVGAVV ILLFVYRDVE YSNELWWQFE FTAEAPRGLR ALLGITIISS AIAIFSLLRP ATFRPEPATE EALTRAVEIV RKQGNADANL VRMGDKSIMF SEKGDAFIMY GRQGRSWIAL FDPVGDHGAV QELVWRFVEA ARAAGCRAVF YQISPALLSH CADAGLRAFK LGELAVADLR TFEMKGGKWA NLRQTASRAQ RDGLEFAVVE PENVPDIIDE LAAVSTAWLE HHNAKEKGFS LGSFDPDYVS AQPVGILKKD GKIVAFANIL VTESKEEGTI DLMRFSPDAP KGSMDFLFVQ IMEYLRNQGF THFNLGMAPL SGMSKREAAP VWDRIGSTVF EHGERFYNFK GLRAFKSKFH PHWQPRYLAV SGGGNPMIAL MDATFLIGGG LKGVVRK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: a waiting time of 10 s and a blotting time of 4 s | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: waiting time of 10 s and blotting time of 4 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Details | CDS mode |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 3159 / Average exposure time: 5.0 sec. / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)