+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Rhizobium etli MprF | |||||||||
 Map data Map data | 3.0 A "Best map" by EMDA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | flippase / aminoacyl-tRNA / phosphatidylglycerol / MEMBRANE PROTEIN | |||||||||
| Function / homology | : / Phosphatidylglycerol lysyltransferase, C-terminal / Phosphatidylglycerol lysyltransferase, C-terminal / aminoacyltransferase activity / phospholipid homeostasis / Acyl-CoA N-acyltransferase / plasma membrane / Hypothetical conserved protein Function and homology information Function and homology information | |||||||||
| Biological species |  Rhizobium etli CFN 42 (bacteria) / Rhizobium etli CFN 42 (bacteria) /  Rhizobium etli (strain CFN 42 / ATCC 51251) (bacteria) Rhizobium etli (strain CFN 42 / ATCC 51251) (bacteria) | |||||||||
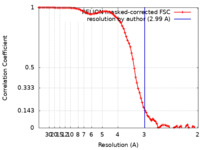
| Method | single particle reconstruction / cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Nishimura M / Hirano H | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Rhizobium etli MprF Authors: Nishimura M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31445.map.gz emd_31445.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31445-v30.xml emd-31445-v30.xml emd-31445.xml emd-31445.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31445_fsc.xml emd_31445_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_31445.png emd_31445.png | 159 KB | ||
| Masks |  emd_31445_msk_1.map emd_31445_msk_1.map | 27.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31445.cif.gz emd-31445.cif.gz | 6.6 KB | ||
| Others |  emd_31445_half_map_1.map.gz emd_31445_half_map_1.map.gz emd_31445_half_map_2.map.gz emd_31445_half_map_2.map.gz | 23.2 MB 23.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31445 http://ftp.pdbj.org/pub/emdb/structures/EMD-31445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31445 | HTTPS FTP |
-Validation report
| Summary document |  emd_31445_validation.pdf.gz emd_31445_validation.pdf.gz | 724.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31445_full_validation.pdf.gz emd_31445_full_validation.pdf.gz | 723.8 KB | Display | |
| Data in XML |  emd_31445_validation.xml.gz emd_31445_validation.xml.gz | 14.4 KB | Display | |
| Data in CIF |  emd_31445_validation.cif.gz emd_31445_validation.cif.gz | 18.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31445 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31445 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31445 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31445 | HTTPS FTP |
-Related structure data
| Related structure data |  7f47MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31445.map.gz / Format: CCP4 / Size: 27.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31445.map.gz / Format: CCP4 / Size: 27.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3.0 A "Best map" by EMDA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.996 Å | ||||||||||||||||||||||||||||||||||||
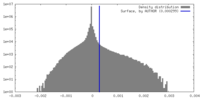
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31445_msk_1.map emd_31445_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
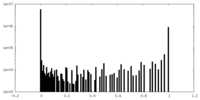
| Density Histograms |
-Half map: #1
| File | emd_31445_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
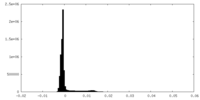
| Density Histograms |
-Half map: #2
| File | emd_31445_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
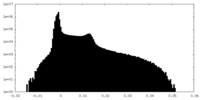
| Density Histograms |
- Sample components
Sample components
-Entire : MprF
| Entire | Name: MprF |
|---|---|
| Components |
|
-Supramolecule #1: MprF
| Supramolecule | Name: MprF / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Rhizobium etli CFN 42 (bacteria) Rhizobium etli CFN 42 (bacteria) |
| Molecular weight | Theoretical: 187 KDa |
-Macromolecule #1: Hypothetical conserved protein
| Macromolecule | Name: Hypothetical conserved protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhizobium etli (strain CFN 42 / ATCC 51251) (bacteria) Rhizobium etli (strain CFN 42 / ATCC 51251) (bacteria) |
| Molecular weight | Theoretical: 93.918508 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGHGNLEEM EATDGFSFRT LFRRYRTPLT AAATLIVFCL VGYAIMQLTN EVRYDDVVAA LAATGPSAIL LALFFTALSF LSLVFYDLN AIEYIGKKLP FPHVALTAFS AYAVGNTAGF GALSGGAIRY RAYTRLGLSP EDIGRIIAFV TLSFGLGLAA V ASIALIII ...String: MSGHGNLEEM EATDGFSFRT LFRRYRTPLT AAATLIVFCL VGYAIMQLTN EVRYDDVVAA LAATGPSAIL LALFFTALSF LSLVFYDLN AIEYIGKKLP FPHVALTAFS AYAVGNTAGF GALSGGAIRY RAYTRLGLSP EDIGRIIAFV TLSFGLGLAA V ASIALIII ASEIGPLIGV SPFLLRLIAG SIIAILGAVM IIGREGRVLN FGAVAIRLPD SRTWSRQFLV TAFDIAASAS VL YVLLPQT AIGWPVFLAV YAIAVGLGVL SHVPAGLGVF ETVIIASLGS AVNIDAVLGS LVLYRLIYHV LPLLIAVLAV SAA ELRRFV DHPAASSVRR IGGRLMPQLL STLALLLGVM LVFSSVTPTP DQNLEFLSNY LPLPMVEGAH FLSSLLGLAL VVAA RGLGQ RLDGAWWVAV FSAVAALTLS LLKAIALVEA AFLAFLIFGL FVSRRLFTRH ASLLNQAMTA SWLMAIAVIV VGAVV ILLF VYRDVEYSNE LWWQFEFTAE APRGLRALLG ITIISSAIAI FSLLRPATFR PEPATEEALT RAVEIVRKQG NADANL VRM GDKSIMFSEK GDAFIMYGRQ GRSWIALFDP VGDHGAVQEL VWRFVEAARA AGCRAVFYQI SPALLSHCAD AGLRAFK LG ELAVADLRTF EMKGGKWANL RQTASRAQRD GLEFAVVEPE NVPDIIDELA AVSTAWLEHH NAKEKGFSLG SFDPDYVS A QPVGILKKDG KIVAFANILV TESKEEGTID LMRFSPDAPK GSMDFLFVQI MEYLRNQGFT HFNLGMAPLS GMSKREAAP VWDRIGSTVF EHGERFYNFK GLRAFKSKFH PHWQPRYLAV SGGGNPMIAL MDATFLIGGG LKGVVRK UniProtKB: Hypothetical conserved protein |
-Macromolecule #2: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]o...
| Macromolecule | Name: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate type: ligand / ID: 2 / Number of copies: 1 / Formula: PGW |
|---|---|
| Molecular weight | Theoretical: 749.007 Da |
-Macromolecule #3: [(2R)-1-[[(2R)-3-[(2S)-2,6-bis(azanyl)hexanoyl]oxy-2-oxidanyl-pro...
| Macromolecule | Name: [(2R)-1-[[(2R)-3-[(2S)-2,6-bis(azanyl)hexanoyl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (E)-octadec-9-enoate type: ligand / ID: 3 / Number of copies: 1 / Formula: 1K1 |
|---|---|
| Molecular weight | Theoretical: 877.179 Da |
| Chemical component information |  ChemComp-1K1: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: a waiting time of 10 s and a blotting time of 4 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Details | CDS mode |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 3159 / Average exposure time: 5.0 sec. / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)