[English] 日本語
 Yorodumi
Yorodumi- EMDB-31342: Cryo-EM structure of cenerimod -bound Sphingosine-1-phosphate rec... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31342 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of cenerimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Homo sapiens / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationadenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin ...adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / synapse / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.07 Å | |||||||||
 Authors Authors | Yuan Y / Jia GW | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2021 Journal: Cell Res / Year: 2021Title: Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition. Authors: Yuan Yuan / Guowen Jia / Chao Wu / Wei Wang / Lin Cheng / Qian Li / Ziyan Li / Kaidong Luo / Shengyong Yang / Wei Yan / Zhaoming Su / Zhenhua Shao /  Abstract: Sphingosine-1-phosphate (S1P) is an important bioactive lipid molecule in cell membrane metabolism and binds to G protein-coupled S1P receptors (S1PRs) to regulate embryonic development, ...Sphingosine-1-phosphate (S1P) is an important bioactive lipid molecule in cell membrane metabolism and binds to G protein-coupled S1P receptors (S1PRs) to regulate embryonic development, physiological homeostasis, and pathogenic processes in various organs. S1PRs are lipid-sensing receptors and are therapeutic targets for drug development, including potential treatment of COVID-19. Herein, we present five cryo-electron microscopy structures of S1PRs bound to diverse drug agonists and the heterotrimeric Gi protein. Our structural and functional assays demonstrate the different binding modes of chemically distinct agonists of S1PRs, reveal the mechanical switch that activates these receptors, and provide a framework for understanding ligand selectivity and G protein coupling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31342.map.gz emd_31342.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31342-v30.xml emd-31342-v30.xml emd-31342.xml emd-31342.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31342.png emd_31342.png | 37.5 KB | ||
| Filedesc metadata |  emd-31342.cif.gz emd-31342.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31342 http://ftp.pdbj.org/pub/emdb/structures/EMD-31342 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31342 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31342 | HTTPS FTP |
-Validation report
| Summary document |  emd_31342_validation.pdf.gz emd_31342_validation.pdf.gz | 474.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31342_full_validation.pdf.gz emd_31342_full_validation.pdf.gz | 474.2 KB | Display | |
| Data in XML |  emd_31342_validation.xml.gz emd_31342_validation.xml.gz | 6.3 KB | Display | |
| Data in CIF |  emd_31342_validation.cif.gz emd_31342_validation.cif.gz | 7.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31342 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31342 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31342 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31342 | HTTPS FTP |
-Related structure data
| Related structure data |  7evzMC  7evyC  7ew0C  7ew1C  7ew7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31342.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31342.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Sphingosine-1-phosphate receptor 1 complexed with Gi protein with...
| Entire | Name: Sphingosine-1-phosphate receptor 1 complexed with Gi protein with bound cenerimod |
|---|---|
| Components |
|
-Supramolecule #1: Sphingosine-1-phosphate receptor 1 complexed with Gi protein with...
| Supramolecule | Name: Sphingosine-1-phosphate receptor 1 complexed with Gi protein with bound cenerimod type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.141793 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHLEVLF QGPGSSGSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSA SQDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT G YLSCCRFL ...String: MHHHHLEVLF QGPGSSGSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSA SQDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT G YLSCCRFL DDNQIVTSSG DTTCALWDIE TGQQTTTFTG HTGDVMSLSL APDTRLFVSG ACDASAKLWD VREGMCRQTF TG HESDINA ICFFPNGNAF ATGSDDATCR LFDLRADQEL MTYSHDNIIC GITSVSFSKS GRLLLAGYDD FNCNVWDALK ADR AGVLAG HDNRVSCLGV TDDGMAVATG SWDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Sphingosine 1-phosphate receptor 1
| Macromolecule | Name: Sphingosine 1-phosphate receptor 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.27375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDANIFEML RIDEGLRLKI YKNTEGYYTI GIGHLLTKSP SLNAAKSELD KAIGRNTNGV ITKDEAEKL FNQDVDAAVR GILRNAKLKP VYDSLDAVRR AALINMVFQM GETGVAGFTN SLRMLQQKRW DEAAVNLAKS R WYNQTPNR ...String: MKTIIALSYI FCLVFADYKD DDDANIFEML RIDEGLRLKI YKNTEGYYTI GIGHLLTKSP SLNAAKSELD KAIGRNTNGV ITKDEAEKL FNQDVDAAVR GILRNAKLKP VYDSLDAVRR AALINMVFQM GETGVAGFTN SLRMLQQKRW DEAAVNLAKS R WYNQTPNR AKRVITTFRT GTWDAYMGPT SVPLVKAHRS SVSDYVNYDI IVRHYNYTGK LNISADKENS IKLTSVVFIL IC CFIILEN IFVLLTIWKT KKFHRPMYYF IGNLALSDLL AGVAYTANLL LSGATTYKLT PAQWFLREGS MFVALSASVF SLL AIAIER YITMLKMKLH NGSNNFRLFL LISACWVISL ILGGLPIMGW NCISALSSCS TVLPLYHKHY ILFCTTVFTL LLLS IVILY CRIYSLVRTR SRRLTFRKNI SKASRSSEKS LALLKTVIIV LSVFIACWAP LFILLLLDVG CKVKTCDILF RAEYF LVLA VLNSGTNPII YTLTNKEMRR AFIRIMSCCK CPSGDSAHHH HHHHHHH |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.636793 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAENLYFQ GHHHHHHHH |
-Macromolecule #6: (2~{S})-3-[4-[5-(2-cyclopentyl-6-methoxy-pyridin-4-yl)-1,2,4-oxad...
| Macromolecule | Name: (2~{S})-3-[4-[5-(2-cyclopentyl-6-methoxy-pyridin-4-yl)-1,2,4-oxadiazol-3-yl]-2-ethyl-6-methyl-phenoxy]propane-1,2-diol type: ligand / ID: 6 / Number of copies: 1 / Formula: JER |
|---|---|
| Molecular weight | Theoretical: 453.531 Da |
| Chemical component information | 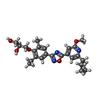 ChemComp-JER: |
-Macromolecule #7: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 7 / Number of copies: 4 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)