+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of VCCN1 in detergent | |||||||||
 Map data Map data | FSC weighted, sharpened and masked map by PostProcess | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / Thylakoid / Photosynthesis / Bestrophin family / MEMBRANE PROTEIN | |||||||||
| Function / homology | UPF0187 protein At3g61320-like / regulation of photosynthesis, light reaction / Bacterial/Fungal Bestrophin protein / UPF0187 family / voltage-gated chloride channel activity / membrane / Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
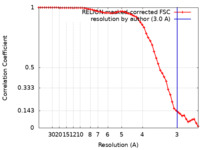
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Hagino T / Kato T / Kasuya G / Kobayashi K / Kusakizako T / Yamashita K / Nishizawa T / Nureki O | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1. Authors: Tatsuya Hagino / Takafumi Kato / Go Kasuya / Kan Kobayashi / Tsukasa Kusakizako / Shin Hamamoto / Tomoaki Sobajima / Yuichiro Fujiwara / Keitaro Yamashita / Hisashi Kawasaki / Andrés D ...Authors: Tatsuya Hagino / Takafumi Kato / Go Kasuya / Kan Kobayashi / Tsukasa Kusakizako / Shin Hamamoto / Tomoaki Sobajima / Yuichiro Fujiwara / Keitaro Yamashita / Hisashi Kawasaki / Andrés D Maturana / Tomohiro Nishizawa / Osamu Nureki /   Abstract: In the light reaction of plant photosynthesis, modulation of electron transport chain reactions is important to maintain the efficiency of photosynthesis under a broad range of light intensities. ...In the light reaction of plant photosynthesis, modulation of electron transport chain reactions is important to maintain the efficiency of photosynthesis under a broad range of light intensities. VCCN1 was recently identified as a voltage-gated chloride channel residing in the thylakoid membrane, where it plays a key role in photoreaction tuning to avoid the generation of reactive oxygen species (ROS). Here, we present the cryo-EM structures of Malus domestica VCCN1 (MdVCCN1) in nanodiscs and detergent at 2.7 Å and 3.0 Å resolutions, respectively, and the structure-based electrophysiological analyses. VCCN1 structurally resembles its animal homolog, bestrophin, a Ca-gated anion channel. However, unlike bestrophin channels, VCCN1 lacks the Ca-binding motif but instead contains an N-terminal charged helix that is anchored to the lipid membrane through an additional amphipathic helix. Electrophysiological experiments demonstrate that these structural elements are essential for the channel activity, thus revealing the distinct activation mechanism of VCCN1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31165.map.gz emd_31165.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31165-v30.xml emd-31165-v30.xml emd-31165.xml emd-31165.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31165_fsc.xml emd_31165_fsc.xml | 5.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_31165.png emd_31165.png | 60.2 KB | ||
| Masks |  emd_31165_msk_1.map emd_31165_msk_1.map | 8.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31165.cif.gz emd-31165.cif.gz | 5.4 KB | ||
| Others |  emd_31165_half_map_1.map.gz emd_31165_half_map_1.map.gz emd_31165_half_map_2.map.gz emd_31165_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31165 http://ftp.pdbj.org/pub/emdb/structures/EMD-31165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31165 | HTTPS FTP |
-Related structure data
| Related structure data |  7ek1MC  7ek2C  7ek3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11091 (Title: Thylakoid-located voltage-dependent chloride channel VCCN1 EMPIAR-11091 (Title: Thylakoid-located voltage-dependent chloride channel VCCN1Data size: 2.2 TB / Data #1: MdVCCN1 in GDN [micrographs - multiframe] / Data #2: MdVCCN1 in nanodiscs [micrographs - multiframe] Data #3: VCCN1 Y332A in nanodiscs [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31165.map.gz / Format: CCP4 / Size: 8.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31165.map.gz / Format: CCP4 / Size: 8.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FSC weighted, sharpened and masked map by PostProcess | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.30406 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31165_msk_1.map emd_31165_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_31165_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_31165_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Bestrophin-like protein
| Entire | Name: Bestrophin-like protein |
|---|---|
| Components |
|
-Supramolecule #1: Bestrophin-like protein
| Supramolecule | Name: Bestrophin-like protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Bestrophin-like protein
| Macromolecule | Name: Bestrophin-like protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.057738 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPNPSPPSSS SPVQTLISIL RIIPDWSDRT QERGMRQHRT LYDHEKWMHH RSSYRHLRHL LSSLSSRVIL SLIPPVIAFT LVAVVIASY NTAVALDLLP GIFPLLRSSS LPYQLTAPAL ALLLVFRTEA SYSRFEEGRK SWTEVIAGAN DFARQIISSV E TSGDAQLK ...String: MPNPSPPSSS SPVQTLISIL RIIPDWSDRT QERGMRQHRT LYDHEKWMHH RSSYRHLRHL LSSLSSRVIL SLIPPVIAFT LVAVVIASY NTAVALDLLP GIFPLLRSSS LPYQLTAPAL ALLLVFRTEA SYSRFEEGRK SWTEVIAGAN DFARQIISSV E TSGDAQLK KALLQYIVAF PVALKCHVIY GSDIARDLQN LLEVDDLLVV LNSKHRPGCI IQFISRSLQL LKLEESRRIM LQ SKISCFH EGIGICEQLI GTPIPLSYTR LTSRFLVLWH LTLPIILWDD CHWIVVPATF ISAASLFCIE QVGVLIEEPF PML ALDDLC NSVRNNVQEA LASEKLIRAR LAAKGRIQSE QQFQNGQPRP ENLYFQ UniProtKB: Uncharacterized protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1161 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)