[English] 日本語
 Yorodumi
Yorodumi- EMDB-30977: SARS-CoV-2 spike in complex with the Ab4 neutralizing antibody (S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30977 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike in complex with the Ab4 neutralizing antibody (State 3) | |||||||||
 Map data Map data | Spike-Ab4 Complex (State 3) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Spike / SARS-CoV-2 / Antibody / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
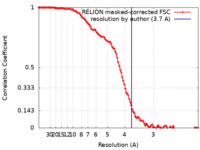
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Liu C | |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2021 Journal: Cell Discov / Year: 2021Title: Three epitope-distinct human antibodies from RenMab mice neutralize SARS-CoV-2 and cooperatively minimize the escape of mutants. Authors: Jianhui Nie / Jingshu Xie / Shuo Liu / Jiajing Wu / Chuan Liu / Jianhui Li / Yacui Liu / Meiyu Wang / Huizhen Zhao / Yabo Zhang / Jiawei Yao / Lei Chen / Yuelei Shen / Yi Yang / Hong-Wei ...Authors: Jianhui Nie / Jingshu Xie / Shuo Liu / Jiajing Wu / Chuan Liu / Jianhui Li / Yacui Liu / Meiyu Wang / Huizhen Zhao / Yabo Zhang / Jiawei Yao / Lei Chen / Yuelei Shen / Yi Yang / Hong-Wei Wang / Youchun Wang / Weijin Huang /  Abstract: Coronavirus disease 2019 (COVID-19), a pandemic disease caused by the newly emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused more than 3.8 million deaths to date. ...Coronavirus disease 2019 (COVID-19), a pandemic disease caused by the newly emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused more than 3.8 million deaths to date. Neutralizing antibodies are effective therapeutic measures. However, many naturally occurring mutations at the receptor-binding domain (RBD) have emerged, and some of them can evade existing neutralizing antibodies. Here, we utilized RenMab, a novel mouse carrying the entire human antibody variable region, for neutralizing antibody discovery. We obtained several potent RBD-blocking antibodies and categorized them into four distinct groups by epitope mapping. We determined the involved residues of the epitope of three representative antibodies by cryo-electron microscopy (Cryo-EM) studies. Moreover, we performed neutralizing experiments with 50 variant strains with single or combined mutations and found that the mixing of three epitope-distinct antibodies almost eliminated the mutant escape. Our study provides a sound basis for the rational design of fully human antibody cocktails against SARS-CoV-2 and pre-emergent coronaviral threats. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30977.map.gz emd_30977.map.gz | 13.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30977-v30.xml emd-30977-v30.xml emd-30977.xml emd-30977.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30977_fsc.xml emd_30977_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_30977.png emd_30977.png | 90.1 KB | ||
| Filedesc metadata |  emd-30977.cif.gz emd-30977.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30977 http://ftp.pdbj.org/pub/emdb/structures/EMD-30977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30977 | HTTPS FTP |
-Related structure data
| Related structure data |  7e39MC  7e3bC  7e3cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30977.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30977.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Spike-Ab4 Complex (State 3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
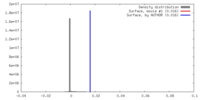
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : complex of spike protein trimer with three Ab4 antibodies
| Entire | Name: complex of spike protein trimer with three Ab4 antibodies |
|---|---|
| Components |
|
-Supramolecule #1: complex of spike protein trimer with three Ab4 antibodies
| Supramolecule | Name: complex of spike protein trimer with three Ab4 antibodies type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: Spike protein S1
| Supramolecule | Name: Spike protein S1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Light Chain of Ab4
| Supramolecule | Name: Light Chain of Ab4 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: Heavy Chain of Ab4
| Supramolecule | Name: Heavy Chain of Ab4 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.776381 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP L QSYGFQPT ...String: TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP L QSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCG UniProtKB: Spike glycoprotein |
-Macromolecule #2: Light Chain of Ab4
| Macromolecule | Name: Light Chain of Ab4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.546252 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NIQMTQSPSA MSASVGDRVT ITCRARQGIS NYLAWFQQKP GKVPKHLIYA ASSLLSGVPS RFSGSGSETE FTLTISSLQP EDFATYYCL QHNSYPYTFG QGTKLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: NIQMTQSPSA MSASVGDRVT ITCRARQGIS NYLAWFQQKP GKVPKHLIYA ASSLLSGVPS RFSGSGSETE FTLTISSLQP EDFATYYCL QHNSYPYTFG QGTKLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: Heavy Chain of Ab4
| Macromolecule | Name: Heavy Chain of Ab4 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.096227 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFIFS SYGMHWVRQA PGKGLEWVAV IWFDGSNKYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARE TVSYGMDVWG QGTTVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV ...String: QVQLVESGGG VVQPGRSLRL SCAASGFIFS SYGMHWVRQA PGKGLEWVAV IWFDGSNKYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARE TVSYGMDVWG QGTTVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKKVEP KSCDKTHTCP PCPAPEAAGG PS VFLFPPK PKDTLMISRT PEVTCVVVDV SHEDPEVKFN WYVDGVEVHN AKTKPREEQY NSTYRVVSVL TVLHQDWLNG KEY KCKVSN KALPAPIEKT ISKAKGQPRE PQVYTLPPSR EEMTKNQVSL TCLVKGFYPS DIAVEWESNG QPENNYKTTP PVLD SDGSF FLYSKLTVDK SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)