[English] 日本語
 Yorodumi
Yorodumi- EMDB-3092: BG505 SOSIP.664 trimer in complex with PGT124 Fab with 32H heavy chain -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3092 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
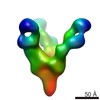
| Title | BG505 SOSIP.664 trimer in complex with PGT124 Fab with 32H heavy chain | |||||||||
 Map data Map data | BG505 SOSIP.664 in complex with a PGT124 precursor Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PGT124 / HIV-1 / broadly neutralizing antibody / Env | |||||||||
| Biological species |   Human Immunodeficiency Virus-1 / Human Immunodeficiency Virus-1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 22.0 Å | |||||||||
 Authors Authors | Lee JH / Garces F / Wilson IA / Ward AB | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2015 Journal: Immunity / Year: 2015Title: Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Authors: Fernando Garces / Jeong Hyun Lee / Natalia de Val / Alba Torrents de la Pena / Leopold Kong / Cristina Puchades / Yuanzi Hua / Robyn L Stanfield / Dennis R Burton / John P Moore / Rogier W ...Authors: Fernando Garces / Jeong Hyun Lee / Natalia de Val / Alba Torrents de la Pena / Leopold Kong / Cristina Puchades / Yuanzi Hua / Robyn L Stanfield / Dennis R Burton / John P Moore / Rogier W Sanders / Andrew B Ward / Ian A Wilson /   Abstract: The high-mannose patch on the HIV-1 envelope (Env) glycoprotein is the epicenter for binding of the potent broadly neutralizing PGT121 family of antibodies, but strategies for generating such ...The high-mannose patch on the HIV-1 envelope (Env) glycoprotein is the epicenter for binding of the potent broadly neutralizing PGT121 family of antibodies, but strategies for generating such antibodies by vaccination have not been defined. We generated structures of inferred antibody intermediates by X-ray crystallography and electron microscopy to elucidate the molecular events that occurred during evolution of this family. Binding analyses revealed that affinity maturation was primarily focused on avoiding, accommodating, or binding the N137 glycan. The overall antibody approach angle to Env was defined very early in the maturation process, yet some variation evolved in the PGT121 family branches that led to differences in glycan specificities in their respective epitopes. Furthermore, we determined a crystal structure of the recombinant BG505 SOSIP.664 HIV-1 trimer with a PGT121 family member at 3.0 Å that, in concert with these antibody intermediate structures, provides insights to advance design of HIV vaccine candidates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3092.map.gz emd_3092.map.gz | 14 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3092-v30.xml emd-3092-v30.xml emd-3092.xml emd-3092.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd-3092_13588.png emd-3092_13588.png | 354.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3092 http://ftp.pdbj.org/pub/emdb/structures/EMD-3092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3092 | HTTPS FTP |
-Validation report
| Summary document |  emd_3092_validation.pdf.gz emd_3092_validation.pdf.gz | 202.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3092_full_validation.pdf.gz emd_3092_full_validation.pdf.gz | 201.9 KB | Display | |
| Data in XML |  emd_3092_validation.xml.gz emd_3092_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3092 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3092 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3092 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3092 | HTTPS FTP |
-Related structure data
| Related structure data |  3093C  6379C  6380C  5cexC  5ceyC  5cezC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3092.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3092.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BG505 SOSIP.664 in complex with a PGT124 precursor Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PGT124 32H+3L Fab in complex with BG505 SOSIP.664
| Entire | Name: PGT124 32H+3L Fab in complex with BG505 SOSIP.664 |
|---|---|
| Components |
|
-Supramolecule #1000: PGT124 32H+3L Fab in complex with BG505 SOSIP.664
| Supramolecule | Name: PGT124 32H+3L Fab in complex with BG505 SOSIP.664 / type: sample / ID: 1000 / Oligomeric state: 3 Fabs bind one SOSIP trimer / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 570 MDa |
-Macromolecule #1: BG505 SOSIP.664 Env trimer
| Macromolecule | Name: BG505 SOSIP.664 Env trimer / type: protein_or_peptide / ID: 1 / Name.synonym: BG505 SOSIP.664 / Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human Immunodeficiency Virus-1 / synonym: HIV-1 Human Immunodeficiency Virus-1 / synonym: HIV-1 |
| Molecular weight | Theoretical: 420 MDa |
| Recombinant expression | Organism: Mammalian (mammals) / Recombinant cell: HEK293S |
-Macromolecule #2: PGT124 Fab with 32H heavy chain
| Macromolecule | Name: PGT124 Fab with 32H heavy chain / type: protein_or_peptide / ID: 2 / Name.synonym: PGT124 32H-3L Details: One light and one heavy chain forms the Fab heterodimer Number of copies: 3 / Oligomeric state: Heterodimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM Tris, 150 mM NaCl |
| Staining | Type: NEGATIVE Details: Grids adsorbed with protein then stained with 2% w/v uranyl formate |
| Grid | Details: 400 mesh Cu grid with carbon support, plasma cleaned |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Temperature | Average: 298 K |
| Alignment procedure | Legacy - Astigmatism: Corrected at 52,000x mag |
| Date | Jun 14, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 460 / Average electron dose: 25 e/Å2 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus min: 0.7 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: OTHER / Software - Name: Sparx / Number images used: 8719 |
|---|
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















