[English] 日本語
 Yorodumi
Yorodumi- EMDB-30914: A refined cryo-EM structure of an Escherichia coli RNAP-promoter ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30914 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A refined cryo-EM structure of an Escherichia coli RNAP-promoter open complex (RPo) with SspA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial RNA polymerase / Complex / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationsigma factor antagonist complex / response to starvation / response to stress / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility ...sigma factor antagonist complex / response to starvation / response to stress / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / regulation of DNA-templated transcription elongation / transcription elongation factor complex / transcription antitermination / cell motility / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / response to heat / intracellular iron ion homeostasis / protein dimerization activity / response to antibiotic / negative regulation of DNA-templated transcription / DNA-templated transcription / positive regulation of DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.68 Å | |||||||||
 Authors Authors | Lin W | |||||||||
 Citation Citation |  Journal: Biochem Biophys Res Commun / Year: 2021 Journal: Biochem Biophys Res Commun / Year: 2021Title: A unique binding between SspA and RNAP βNTH across low-GC Gram-negative bacteria facilitates SspA-mediated transcription regulation. Authors: Fulin Wang / Yu Feng / Zhuo Shang / Wei Lin /  Abstract: Stringent starvation protein A (SspA) involved in nucleotide metabolism, acid tolerance and virulence of bacteria has been demonstrated to function as a transcription factor to regulate σ-dependent ...Stringent starvation protein A (SspA) involved in nucleotide metabolism, acid tolerance and virulence of bacteria has been demonstrated to function as a transcription factor to regulate σ-dependent gene transcription through interacting with σ region 4 and the zinc binding domain (ZBD) of E. coli RNA polymerase (EcoRNAP) β' subunit simultaneously. Despite extensive biochemical and structural analyses were reported recently, the interactions of SspA with RNAP are not comprehensively understood. Here, we reprocessed our previous cryo-EM dataset of EcoRNAP-promoter open complex with SspA (SspA-RPo) and obtained a significantly improved density map. Unexpectedly, the new map showed that SspA interacts with both N-terminal helix of β' subunit (β'ΝΤΗ) and ω subunit, which contributes to stabilize the SspA-EcoRNAP σ holoenzyme complex. Sequence alignments and phylogenetic tree analyses of N-terminal sequences of β' subunit from different classes of bacteria revealed that β'ΝΤΗ is highly conserved and exclusively found in low-GC-content Gram-negative bacteria that harbor SspA, implying a co-evolution of β'ΝΤΗ and SspA. The transcription assays of wild-type SspA and its mutants demonstrated the interaction between SspA and β'ΝΤΗ facilitates the transcription regulation of SspA. Together, our results provide a more comprehensive insight into the interactions between SspA and RNAP and their roles in bacterial transcription regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30914.map.gz emd_30914.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30914-v30.xml emd-30914-v30.xml emd-30914.xml emd-30914.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30914.png emd_30914.png | 123.6 KB | ||
| Filedesc metadata |  emd-30914.cif.gz emd-30914.cif.gz | 8.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30914 http://ftp.pdbj.org/pub/emdb/structures/EMD-30914 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30914 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30914 | HTTPS FTP |
-Validation report
| Summary document |  emd_30914_validation.pdf.gz emd_30914_validation.pdf.gz | 598.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30914_full_validation.pdf.gz emd_30914_full_validation.pdf.gz | 598.2 KB | Display | |
| Data in XML |  emd_30914_validation.xml.gz emd_30914_validation.xml.gz | 5.7 KB | Display | |
| Data in CIF |  emd_30914_validation.cif.gz emd_30914_validation.cif.gz | 6.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30914 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30914 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30914 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30914 | HTTPS FTP |
-Related structure data
| Related structure data |  7dy6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30914.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30914.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.307 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
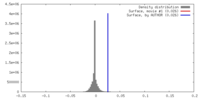
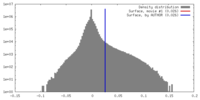
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : an Escherichia coli RNAP-promoter open complex (RPo) with SspA
+Supramolecule #1: an Escherichia coli RNAP-promoter open complex (RPo) with SspA
+Macromolecule #1: DNA (63-MER)
+Macromolecule #7: DNA (63-MER)
+Macromolecule #2: Stringent starvation protein A
+Macromolecule #3: DNA-directed RNA polymerase subunit alpha
+Macromolecule #4: DNA-directed RNA polymerase subunit beta
+Macromolecule #5: DNA-directed RNA polymerase subunit beta'
+Macromolecule #6: RNA polymerase sigma factor RpoD
+Macromolecule #8: DNA-directed RNA polymerase subunit omega
+Macromolecule #9: MAGNESIUM ION
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Grid | Material: COPPER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)