[English] 日本語
 Yorodumi
Yorodumi- EMDB-30790: Cryo-EM structure of the human ABCB6 (coproporphyrin III-bound) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human ABCB6 (coproporphyrin III-bound) | |||||||||||||||
 Map data Map data | generated from non-uniform refinement in cryosparc | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ABCB6 / heme transporter / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDefective ABCB6 causes MCOPCB7 / cellular detoxification of cadmium ion / Mitochondrial ABC transporters / tetrapyrrole metabolic process / ABC-type heme transporter / porphyrin-containing compound metabolic process / tetrapyrrole binding / heme metabolic process / porphyrin-containing compound biosynthetic process / heme transport ...Defective ABCB6 causes MCOPCB7 / cellular detoxification of cadmium ion / Mitochondrial ABC transporters / tetrapyrrole metabolic process / ABC-type heme transporter / porphyrin-containing compound metabolic process / tetrapyrrole binding / heme metabolic process / porphyrin-containing compound biosynthetic process / heme transport / melanosome assembly / ABC-type heme transporter activity / heme transmembrane transport / melanosome membrane / multivesicular body membrane / mitochondrial envelope / endolysosome membrane / vacuolar membrane / skin development / efflux transmembrane transporter activity / intracellular copper ion homeostasis / ABC-type transporter activity / ATP-binding cassette (ABC) transporter complex / brain development / transmembrane transport / early endosome membrane / mitochondrial outer membrane / intracellular iron ion homeostasis / endosome / Golgi membrane / lysosomal membrane / heme binding / endoplasmic reticulum membrane / endoplasmic reticulum / Golgi apparatus / ATP hydrolysis activity / mitochondrion / extracellular exosome / nucleoplasm / ATP binding / plasma membrane / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||
 Authors Authors | Kim S / Lee SS | |||||||||||||||
| Funding support |  Korea, Republic Of, 4 items Korea, Republic Of, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cells / Year: 2022 Journal: Mol Cells / Year: 2022Title: Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6. Authors: Songwon Kim / Sang Soo Lee / Jun Gyou Park / Ji Won Kim / Seulgi Ju / Seung Hun Choi / Subin Kim / Na Jin Kim / Semi Hong / Jin Young Kang / Mi Sun Jin /  Abstract: Human ABCB6 is an ATP-binding cassette transporter that regulates heme biosynthesis by translocating various porphyrins from the cytoplasm into the mitochondria. Here we report the cryo-electron ...Human ABCB6 is an ATP-binding cassette transporter that regulates heme biosynthesis by translocating various porphyrins from the cytoplasm into the mitochondria. Here we report the cryo-electron microscopy (cryo-EM) structures of human ABCB6 with its substrates, coproporphyrin III (CPIII) and hemin, at 3.5 and 3.7 Å resolution, respectively. Metalfree porphyrin CPIII binds to ABCB6 within the central cavity, where its propionic acids form hydrogen bonds with the highly conserved Y550. The resulting structure has an overall fold similar to the inward-facing apo structure, but the two nucleotide-binding domains (NBDs) are slightly closer to each other. In contrast, when ABCB6 binds a metal-centered porphyrin hemin in complex with two glutathione molecules (1 hemin: 2 glutathione), the two NBDs end up much closer together, aligning them to bind and hydrolyze ATP more efficiently. In our structures, a glycine-rich and highly flexible "bulge" loop on TM helix 7 undergoes significant conformational changes associated with substrate binding. Our findings suggest that ABCB6 utilizes at least two distinct mechanisms to fine-tune substrate specificity and transport efficiency. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30790.map.gz emd_30790.map.gz | 141.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30790-v30.xml emd-30790-v30.xml emd-30790.xml emd-30790.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30790.png emd_30790.png | 30.6 KB | ||
| Filedesc metadata |  emd-30790.cif.gz emd-30790.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30790 http://ftp.pdbj.org/pub/emdb/structures/EMD-30790 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30790 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30790 | HTTPS FTP |
-Validation report
| Summary document |  emd_30790_validation.pdf.gz emd_30790_validation.pdf.gz | 463.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30790_full_validation.pdf.gz emd_30790_full_validation.pdf.gz | 463.2 KB | Display | |
| Data in XML |  emd_30790_validation.xml.gz emd_30790_validation.xml.gz | 6.8 KB | Display | |
| Data in CIF |  emd_30790_validation.cif.gz emd_30790_validation.cif.gz | 7.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30790 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30790 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30790 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30790 | HTTPS FTP |
-Related structure data
| Related structure data |  7dnyMC  7dnzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30790.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30790.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | generated from non-uniform refinement in cryosparc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.656 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : ABCB6
| Entire | Name: ABCB6 |
|---|---|
| Components |
|
-Supramolecule #1: ABCB6
| Supramolecule | Name: ABCB6 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATP-binding cassette sub-family B member 6, mitochondrial
| Macromolecule | Name: ATP-binding cassette sub-family B member 6, mitochondrial type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.974172 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVTVGNYCEA EGPVGPAWMQ DGLSPCFFFT LVPSTRMALG TLALVLALPC RRRERPAGAD SLSWGAGPRI SPYVLQLLLA TLQAALPLA GLAGRVGTAR GAPLPSYLLL ASVLESLAGA CGLWLLVVER SQARQRLAMG IWIKFRHSPG LLLLWTVAFA A ENLALVSW ...String: MVTVGNYCEA EGPVGPAWMQ DGLSPCFFFT LVPSTRMALG TLALVLALPC RRRERPAGAD SLSWGAGPRI SPYVLQLLLA TLQAALPLA GLAGRVGTAR GAPLPSYLLL ASVLESLAGA CGLWLLVVER SQARQRLAMG IWIKFRHSPG LLLLWTVAFA A ENLALVSW NSPQWWWARA DLGQQVQFSL WVLRYVVSGG LFVLGLWAPG LRPQSYTLQV HEEDQDVERS QVRSAAQQST WR DFGRKLR LLSGYLWPRG SPALQLVVLI CLGLMGLERA LNVLVPIFYR NIVNLLTEKA PWNSLAWTVT SYVFLKFLQG GGT GSTGFV SNLRTFLWIR VQQFTSRRVE LLIFSHLHEL SLRWHLGRRT GEVLRIADRG TSSVTGLLSY LVFNVIPTLA DIII GIIYF SMFFNAWFGL IVFLCMSLYL TLTIVVTEWR TKFRRAMNTQ ENATRARAVD SLLNFETVKY YNAESYEVER YREAI IKYQ GLEWKSSASL VLLNQTQNLV IGLGLLAGSL LCAYFVTEQK LQVGDYVLFG TYIIQLYMPL NWFGTYYRMI QTNFID MEN MFDLLKEETE VKDLPGAGPL RFQKGRIEFE NVHFSYADGR ETLQDVSFTV MPGQTLALVG PSGAGKSTIL RLLFRFY DI SSGCIRIDGQ DISQVTQASL RSHIGVVPQD TVLFNDTIAD NIRYGRVTAG NDEVEAAAQA AGIHDAIMAF PEGYRTQV G ERGLKLSGGE KQRVAIARTI LKAPGIILLD EATSALDTSN ERAIQASLAK VCANRTTIVV AHRLSTVVNA DQILVIKDG CIVERGRHEA LLSRGGVYAD MWQLQQGQEE TSEDTKPQTM ER UniProtKB: ATP-binding cassette sub-family B member 6 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 5 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: coproporphyrin III
| Macromolecule | Name: coproporphyrin III / type: ligand / ID: 3 / Number of copies: 1 / Formula: HT9 |
|---|---|
| Molecular weight | Theoretical: 654.709 Da |
| Chemical component information | 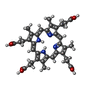 ChemComp-HT9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7 / Component - Concentration: 200.0 mM / Component - Formula: NaCl / Component - Name: sodium chloride |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7dny: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















