+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30665 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of histamine H1 receptor Gq complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHistamine receptors / histamine receptor activity / regulation of vascular permeability / cellular response to histamine / positive regulation of vasoconstriction / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / regulation of synaptic plasticity / visual learning / memory / Olfactory Signaling Pathway ...Histamine receptors / histamine receptor activity / regulation of vascular permeability / cellular response to histamine / positive regulation of vasoconstriction / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / regulation of synaptic plasticity / visual learning / memory / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / chemical synaptic transmission / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / inflammatory response / lysosomal membrane / GTPase activity / synapse / dendrite / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
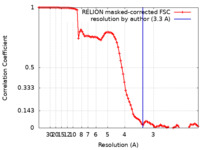
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | He Y / Xia R | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM structure of the human histamine H receptor/G complex. Authors: Ruixue Xia / Na Wang / Zhenmei Xu / Yang Lu / Jing Song / Anqi Zhang / Changyou Guo / Yuanzheng He /  Abstract: Histamine receptors play important roles in various pathophysiological conditions and are effective targets for anti-allergy treatment, however the mechanism of receptor activation remain elusive. ...Histamine receptors play important roles in various pathophysiological conditions and are effective targets for anti-allergy treatment, however the mechanism of receptor activation remain elusive. Here, we present the cryo-electron microscopy (cryo-EM) structure of the human HR in complex with a G protein in an active conformation via a NanoBiT tethering strategy. The structure reveals that histamine activates receptor via interacting with the key residues of both transmembrane domain 3 (TM3) and TM6 to squash the binding pocket on the extracellular side and to open the cavity on the intracellular side for G engagement in a model of "squash to activate and expand to deactivate". The structure also reveals features for G coupling, including the interaction between intracellular loop 2 (ICL2) and the αN-β junction of G protein. The detailed analysis of our structure will provide a framework for understanding G-protein coupling selectivity and clues for designing novel antihistamines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30665.map.gz emd_30665.map.gz | 33 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30665-v30.xml emd-30665-v30.xml emd-30665.xml emd-30665.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30665_fsc.xml emd_30665_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_30665.png emd_30665.png | 69 KB | ||
| Filedesc metadata |  emd-30665.cif.gz emd-30665.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30665 http://ftp.pdbj.org/pub/emdb/structures/EMD-30665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30665 | HTTPS FTP |
-Validation report
| Summary document |  emd_30665_validation.pdf.gz emd_30665_validation.pdf.gz | 526.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30665_full_validation.pdf.gz emd_30665_full_validation.pdf.gz | 525.9 KB | Display | |
| Data in XML |  emd_30665_validation.xml.gz emd_30665_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_30665_validation.cif.gz emd_30665_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30665 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30665 | HTTPS FTP |
-Related structure data
| Related structure data |  7dflMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30665.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30665.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GPCR/G-protein complex
| Entire | Name: GPCR/G-protein complex |
|---|---|
| Components |
|
-Supramolecule #1: GPCR/G-protein complex
| Supramolecule | Name: GPCR/G-protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 130 kDa/nm |
-Macromolecule #1: Histamine H1 receptor
| Macromolecule | Name: Histamine H1 receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.856035 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLPNSSCLL EDKMCEGNKT TMASPQLMPL VVVLSTICLV TVGLNLLVLY AVRSERKLHT VGNLYIVSLS VADLIVGAVV MPMNILYLL MSKWSLGRPL CLFWLSMDYV ASTASIFSVF ILCIDRYRSV QQPLRYLKYR TKTRASATIL GAWFLSFLWV I PILGWNHF ...String: MSLPNSSCLL EDKMCEGNKT TMASPQLMPL VVVLSTICLV TVGLNLLVLY AVRSERKLHT VGNLYIVSLS VADLIVGAVV MPMNILYLL MSKWSLGRPL CLFWLSMDYV ASTASIFSVF ILCIDRYRSV QQPLRYLKYR TKTRASATIL GAWFLSFLWV I PILGWNHF MQQTSVRRED KCETDFYDVT WFKVMTAIIN FYLPTLLMLW FYAKIYKAVR QHCQHRELIN RSLPSFSEIK LR PENPKGD AKKPGKESPW EVLKRKPKDA GGGSVLKSPS QTPKEMKSPV VFSQEDDREV DKLYCFPLDI VHMQAAAEGS SRD YVAVNR SHGQLKTDEQ GLNTHGASEI SEDQMLGDSQ SFSRTDSDTT TETAPGKGKL RSGSNTGLDY IKFTWKRLRS HSRQ YVSGL HMNRERKAAK QLGFIMAAFI LCWIPYFIFF MVIAFCKNCC NEHLHMFTIW LGYINSTLNP LIYPLCNENF KKTFK RILH IRS UniProtKB: Histamine H1 receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.285734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Guanine nucleotide-binding protein G(q) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(q) subunit alpha / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.090637 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CTLSAEDKAA VERSKMIDRN LREDGEKARR ELKLLLLGTG ESGKSTFIKQ MRIIHGSGYS DEDKRGFTKL VYQNIFTAMQ AMIRAMDTL KIPYKHEHNK AHAQLVREVD VEKVSTFENP YVDAIRSLWS DPGIQECYDR RREYQLSDST KYYLNDLDRI A DSTYLPTQ ...String: CTLSAEDKAA VERSKMIDRN LREDGEKARR ELKLLLLGTG ESGKSTFIKQ MRIIHGSGYS DEDKRGFTKL VYQNIFTAMQ AMIRAMDTL KIPYKHEHNK AHAQLVREVD VEKVSTFENP YVDAIRSLWS DPGIQECYDR RREYQLSDST KYYLNDLDRI A DSTYLPTQ QDVLRVRVPT TGIIEYPFDL QSVIFRMVDV GGQRSERRKW IHCFENVTSI MFLVALSEYD QVLVESDNEN RM EESKALF RTIITYPWFQ NSSVILFLNK KDLLEEKIMY SHLVDYFPEY DGPQRDAQAA REFILKMFVD LNPDSDKILY SHF TCATDT ENIRFVFAAV KDTILQLNLK EYNLV |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.679721 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAA |
-Macromolecule #6: HISTAMINE
| Macromolecule | Name: HISTAMINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: HSM |
|---|---|
| Molecular weight | Theoretical: 111.145 Da |
| Chemical component information | 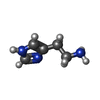 ChemComp-HSM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)