+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30661 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 S-ACE2 complex | |||||||||
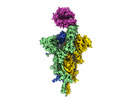
 Map data Map data | S-ACE2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Spike glycoprotein / coronavirus / SARS-CoV-2 virus / receptor ACE2 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy ...positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy / peptidyl-dipeptidase activity / regulation of vasoconstriction / transporter activator activity / Metabolism of Angiotensinogen to Angiotensins / carboxypeptidase activity / angiotensin maturation / viral life cycle / Attachment and Entry / receptor-mediated endocytosis of virus by host cell / metallocarboxypeptidase activity / positive regulation of cardiac muscle contraction / regulation of cytokine production / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / negative regulation of ERK1 and ERK2 cascade / positive regulation of reactive oxygen species metabolic process / metallopeptidase activity / endocytic vesicle membrane / regulation of cell population proliferation / virus receptor activity / regulation of inflammatory response / endopeptidase activity / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Potential therapeutics for SARS / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / cilium / symbiont-mediated suppression of host innate immune response / apical plasma membrane / receptor ligand activity / membrane raft / endocytosis involved in viral entry into host cell / endoplasmic reticulum lumen / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / cell surface / negative regulation of transcription by RNA polymerase II / extracellular space / extracellular exosome / extracellular region / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Cong X / Yao C | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Authors: Cong Xu / Yanxing Wang / Caixuan Liu / Chao Zhang / Wenyu Han / Xiaoyu Hong / Yifan Wang / Qin Hong / Shutian Wang / Qiaoyu Zhao / Yalei Wang / Yong Yang / Kaijian Chen / Wei Zheng / ...Authors: Cong Xu / Yanxing Wang / Caixuan Liu / Chao Zhang / Wenyu Han / Xiaoyu Hong / Yifan Wang / Qin Hong / Shutian Wang / Qiaoyu Zhao / Yalei Wang / Yong Yang / Kaijian Chen / Wei Zheng / Liangliang Kong / Fangfang Wang / Qinyu Zuo / Zhong Huang / Yao Cong /  Abstract: The recent outbreaks of SARS-CoV-2 pose a global health emergency. The SARS-CoV-2 trimeric spike (S) glycoprotein interacts with the human ACE2 receptor to mediate viral entry into host cells. We ...The recent outbreaks of SARS-CoV-2 pose a global health emergency. The SARS-CoV-2 trimeric spike (S) glycoprotein interacts with the human ACE2 receptor to mediate viral entry into host cells. We report the cryo-EM structures of a tightly closed SARS-CoV-2 S trimer with packed fusion peptide and an ACE2-bound S trimer at 2.7- and 3.8-Å resolution, respectively. Accompanying ACE2 binding to the up receptor-binding domain (RBD), the associated ACE2-RBD exhibits continuous swing motions. Notably, the SARS-CoV-2 S trimer appears much more sensitive to the ACE2 receptor than the SARS-CoV S trimer regarding receptor-triggered transformation from the closed prefusion state to the fusion-prone open state, potentially contributing to the superior infectivity of SARS-CoV-2. We defined the RBD T470-T478 loop and Y505 as viral determinants for specific recognition of SARS-CoV-2 RBD by ACE2. Our findings depict the mechanism of ACE2-induced S trimer conformational transitions from the ground prefusion state toward the postfusion state, facilitating development of anti-SARS-CoV-2 vaccines and therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30661.map.gz emd_30661.map.gz | 4.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30661-v30.xml emd-30661-v30.xml emd-30661.xml emd-30661.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30661.png emd_30661.png | 101.3 KB | ||
| Filedesc metadata |  emd-30661.cif.gz emd-30661.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30661 http://ftp.pdbj.org/pub/emdb/structures/EMD-30661 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30661 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30661 | HTTPS FTP |
-Validation report
| Summary document |  emd_30661_validation.pdf.gz emd_30661_validation.pdf.gz | 364.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30661_full_validation.pdf.gz emd_30661_full_validation.pdf.gz | 364.4 KB | Display | |
| Data in XML |  emd_30661_validation.xml.gz emd_30661_validation.xml.gz | 7.2 KB | Display | |
| Data in CIF |  emd_30661_validation.cif.gz emd_30661_validation.cif.gz | 8.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30661 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30661 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30661 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30661 | HTTPS FTP |
-Related structure data
| Related structure data |  7df4MC  7df3C  7dk3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30661.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30661.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | S-ACE2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 spike glycoprotein trimer
| Entire | Name: SARS-CoV-2 spike glycoprotein trimer |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike glycoprotein trimer
| Supramolecule | Name: SARS-CoV-2 spike glycoprotein trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Angiotensin-converting enzyme 2
| Macromolecule | Name: Angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: angiotensin-converting enzyme 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.396492 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHSSALLCCL VLLTGVRAQS TIEEQAKTFL DKFNHEAEDL FYQSSLASWN YNTNITEENV QNMNNAGDKW SAFLKEQSTL AQMYPLQEI QNLTVKLQLQ ALQQNGSSVL SEDKSKRLNT ILNTMSTIYS TGKVCNPDNP QECLLLEPGL NEIMANSLDY N ERLWAWES ...String: MHSSALLCCL VLLTGVRAQS TIEEQAKTFL DKFNHEAEDL FYQSSLASWN YNTNITEENV QNMNNAGDKW SAFLKEQSTL AQMYPLQEI QNLTVKLQLQ ALQQNGSSVL SEDKSKRLNT ILNTMSTIYS TGKVCNPDNP QECLLLEPGL NEIMANSLDY N ERLWAWES WRSEVGKQLR PLYEEYVVLK NEMARANHYE DYGDYWRGDY EVNGVDGYDY SRGQLIEDVE HTFEEIKPLY EH LHAYVRA KLMNAYPSYI SPIGCLPAHL LGDMWGRFWT NLYSLTVPFG QKPNIDVTDA MVDQAWDAQR IFKEAEKFFV SVG LPNMTQ GFWENSMLTD PGNVQKAVCH PTAWDLGKGD FRILMCTKVT MDDFLTAHHE MGHIQYDMAY AAQPFLLRNG ANEG FHEAV GEIMSLSAAT PKHLKSIGLL SPDFQEDNET EINFLLKQAL TIVGTLPFTY MLEKWRWMVF KGEIPKDQWM KKWWE MKRE IVGVVEPVPH DETYCDPASL FHVSNDYSFI RYYTRTLYQF QFQEALCQAA KHEGPLHKCD ISNSTEAGQK LFNMLR LGK SEPWTLALEN VVGAKNMNVR PLLNYFEPLF TWLKDQNKNS FVGWSTDWSP YADHHHHHHH HH UniProtKB: Angiotensin-converting enzyme 2 |
-Macromolecule #2: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.055906 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGAALQIPFA MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STASALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQGSGYIPE APRDGQAYVR KDGEWVLLST FLENLYFQGD YKDDDDKHHH HHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 36 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















