[English] 日本語
 Yorodumi
Yorodumi- EMDB-29915: CryoEM map of a de novo designed octahedral nanocage with program... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map of a de novo designed octahedral nanocage with programmable volume; design cage_O4_34 | |||||||||
 Map data Map data | DeepEMhancer sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | octahedral nanocage / octahedra / nanocage / nanomaterial / computational design / de novo / DE NOVO PROTEIN | |||||||||
| Biological species | synthetic construct (others) | |||||||||
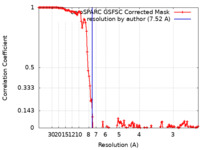
| Method | single particle reconstruction / cryo EM / Resolution: 7.52 Å | |||||||||
 Authors Authors | Kibler RD / Borst AJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Blueprinting expandable nanomaterials with standardized protein building blocks. Abstract: A wooden house frame consists of many different lumber pieces, but because of the regularity of these building blocks, the structure can be designed using straightforward geometrical principles. The ...A wooden house frame consists of many different lumber pieces, but because of the regularity of these building blocks, the structure can be designed using straightforward geometrical principles. The design of multicomponent protein assemblies in comparison has been much more complex, largely due to the irregular shapes of protein structures . Here we describe extendable linear, curved, and angled protein building blocks, as well as inter-block interactions that conform to specified geometric standards; assemblies designed using these blocks inherit their extendability and regular interaction surfaces, enabling them to be expanded or contracted by varying the number of modules, and reinforced with secondary struts. Using X-ray crystallography and electron microscopy, we validate nanomaterial designs ranging from simple polygonal and circular oligomers that can be concentrically nested, up to large polyhedral nanocages and unbounded straight "train track" assemblies with reconfigurable sizes and geometries that can be readily blueprinted. Because of the complexity of protein structures and sequence-structure relationships, it has not been previously possible to build up large protein assemblies by deliberate placement of protein backbones onto a blank 3D canvas; the simplicity and geometric regularity of our design platform now enables construction of protein nanomaterials according to "back of an envelope" architectural blueprints. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29915.map.gz emd_29915.map.gz | 196.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29915-v30.xml emd-29915-v30.xml emd-29915.xml emd-29915.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29915_fsc.xml emd_29915_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_29915.png emd_29915.png | 114.1 KB | ||
| Others |  emd_29915_additional_1.map.gz emd_29915_additional_1.map.gz emd_29915_half_map_1.map.gz emd_29915_half_map_1.map.gz emd_29915_half_map_2.map.gz emd_29915_half_map_2.map.gz | 98.2 MB 192.4 MB 192.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29915 http://ftp.pdbj.org/pub/emdb/structures/EMD-29915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29915 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29915.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29915.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.248 Å | ||||||||||||||||||||||||||||||||||||
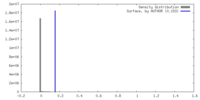
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_29915_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
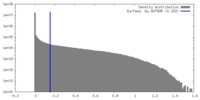
| Density Histograms |
-Half map: Half map A
| File | emd_29915_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
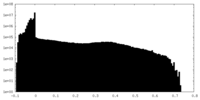
| Density Histograms |
-Half map: Half map B
| File | emd_29915_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
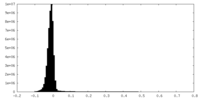
| Density Histograms |
- Sample components
Sample components
-Entire : cage_O4_34
| Entire | Name: cage_O4_34 |
|---|---|
| Components |
|
-Supramolecule #1: cage_O4_34
| Supramolecule | Name: cage_O4_34 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The smallest size of a de novo designed octahedral nanocage with programmable volume |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: cage_O4_34
| Macromolecule | Name: cage_O4_34 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:  |
| Sequence | String: MEEKREIAEF AREMMEEISE RVERYVEDPV LATAVRGRAI VESAAVFAKT IGGDIEKLDE AREEVERAAE EALRRREEGA DVSELVAELI RETSRQIAEI AEATIKATDD PEVLEEISEF AEERSRRLSE YAERHVTNPI LAATVVALAE VLSAVVRARS YGAPEEVGEK ...String: MEEKREIAEF AREMMEEISE RVERYVEDPV LATAVRGRAI VESAAVFAKT IGGDIEKLDE AREEVERAAE EALRRREEGA DVSELVAELI RETSRQIAEI AEATIKATDD PEVLEEISEF AEERSRRLSE YAERHVTNPI LAATVVALAE VLSAVVRARS YGAPEEVGEK AVKEVREASE EALERYKEGA DESELIAEIM EATAEAVGKI AEAAIEATDD PEKRRRIARW AREQMRRLSR RAEELVEDPV LAMTVFARAQ ILAAAVFGKA VGMPEIVSEH ARRMVRLAGE AALALKKEGA DVSRLVAMVG RVTSIAVGMI AKATVSDSPV EITKELIRAF KEITEELIKA GVDGKHLNEA VRLGSEAVNE LVEEAVKEGT PVEEVTESAI KGLEAVTEMA IEAMKAGGDP LEMARIVAEL AEKAIDVVAK MGAPGTYMLA MIMAASVAHR RIVEAAIKAG APGEVLEEAV RLAGRVQVRG VEKAAELGTP KTLVEAAGEL GLATVRRMAE LAIEAGGDRE RMEEIIREVE EAMERVIERI EGGSGGSWGH HHHHHG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 25mM Tris, 300mM NaCl |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS TUNDRA |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Average electron dose: 42.6 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 17.0 µm / Nominal defocus min: 8.0 µm |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)