[English] 日本語
 Yorodumi
Yorodumi- EMDB-29909: Structure of human NDS.3 Fab in complex with influenza virus neur... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) | |||||||||
 Map data Map data | Map post-processed with DeepEMhancer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | neuraminidase / antibody / influenza / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationexo-alpha-sialidase / exo-alpha-sialidase activity / viral budding from plasma membrane / carbohydrate metabolic process / host cell plasma membrane / virion membrane / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Influenza A virus Influenza A virus | |||||||||
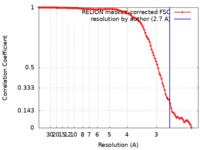
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Tsybovsky Y / Lederhofer J / Kwong PD / Kanekiyo M | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2024 Journal: Immunity / Year: 2024Title: Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase. Authors: Julia Lederhofer / Yaroslav Tsybovsky / Lam Nguyen / Julie E Raab / Adrian Creanga / Tyler Stephens / Rebecca A Gillespie / Hubza Z Syeda / Brian E Fisher / Michelle Skertic / Christina Yap ...Authors: Julia Lederhofer / Yaroslav Tsybovsky / Lam Nguyen / Julie E Raab / Adrian Creanga / Tyler Stephens / Rebecca A Gillespie / Hubza Z Syeda / Brian E Fisher / Michelle Skertic / Christina Yap / Andrew J Schaub / Reda Rawi / Peter D Kwong / Barney S Graham / Adrian B McDermott / Sarah F Andrews / Neil P King / Masaru Kanekiyo /  Abstract: Continuously evolving influenza viruses cause seasonal epidemics and pose global pandemic threats. Although viral neuraminidase (NA) is an effective drug and vaccine target, our understanding of the ...Continuously evolving influenza viruses cause seasonal epidemics and pose global pandemic threats. Although viral neuraminidase (NA) is an effective drug and vaccine target, our understanding of the NA antigenic landscape still remains incomplete. Here, we describe NA-specific human antibodies that target the underside of the NA globular head domain, inhibit viral propagation of a wide range of human H3N2, swine-origin variant H3N2, and H2N2 viruses, and confer both pre- and post-exposure protection against lethal H3N2 infection in mice. Cryo-EM structures of two such antibodies in complex with NA reveal non-overlapping epitopes covering the underside of the NA head. These sites are highly conserved among N2 NAs yet inaccessible unless the NA head tilts or dissociates. Our findings help guide the development of effective countermeasures against ever-changing influenza viruses by identifying hidden conserved sites of vulnerability on the NA underside. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29909.map.gz emd_29909.map.gz | 85.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29909-v30.xml emd-29909-v30.xml emd-29909.xml emd-29909.xml | 28.2 KB 28.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29909_fsc.xml emd_29909_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_29909.png emd_29909.png | 64.1 KB | ||
| Masks |  emd_29909_msk_1.map emd_29909_msk_1.map | 98.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29909.cif.gz emd-29909.cif.gz | 7.8 KB | ||
| Others |  emd_29909_additional_1.map.gz emd_29909_additional_1.map.gz emd_29909_additional_2.map.gz emd_29909_additional_2.map.gz emd_29909_half_map_1.map.gz emd_29909_half_map_1.map.gz emd_29909_half_map_2.map.gz emd_29909_half_map_2.map.gz | 8.7 MB 73.1 MB 73.2 MB 73.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29909 http://ftp.pdbj.org/pub/emdb/structures/EMD-29909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29909 | HTTPS FTP |
-Validation report
| Summary document |  emd_29909_validation.pdf.gz emd_29909_validation.pdf.gz | 756.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29909_full_validation.pdf.gz emd_29909_full_validation.pdf.gz | 756.4 KB | Display | |
| Data in XML |  emd_29909_validation.xml.gz emd_29909_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_29909_validation.cif.gz emd_29909_validation.cif.gz | 23.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29909 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29909 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29909 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29909 | HTTPS FTP |
-Related structure data
| Related structure data |  8gavMC  8gatC  8gauC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29909.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29909.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map post-processed with DeepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29909_msk_1.map emd_29909_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map automatically post-processed with Relion
| File | emd_29909_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map automatically post-processed with Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map before post-processing
| File | emd_29909_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before post-processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_29909_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_29909_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) in com...
| Entire | Name: Influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) in complex with Fab NDS.3 |
|---|---|
| Components |
|
-Supramolecule #1: Influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) in com...
| Supramolecule | Name: Influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) in complex with Fab NDS.3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 380 KDa |
-Macromolecule #1: Neuraminidase
| Macromolecule | Name: Neuraminidase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/Darwin/09/2021 (H3N2) Influenza A virus / Strain: A/Darwin/09/2021 (H3N2) |
| Molecular weight | Theoretical: 51.972344 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEFGLSWIFL AAILKGVQCA DPHHHHHHHH SSSDRFDVDK LKQEIMTEIR REVSKMKQDI IEAIKVELNR GSLVPRGSGG EYRNWSKPQ CGITGFAPFS KDNSIRLSAG GDIWVTREPY VSCDLDKCYQ FALGQGTTLN NVHSNNTVSD RTPYRTLLMN E LGVPFHLG ...String: MEFGLSWIFL AAILKGVQCA DPHHHHHHHH SSSDRFDVDK LKQEIMTEIR REVSKMKQDI IEAIKVELNR GSLVPRGSGG EYRNWSKPQ CGITGFAPFS KDNSIRLSAG GDIWVTREPY VSCDLDKCYQ FALGQGTTLN NVHSNNTVSD RTPYRTLLMN E LGVPFHLG TKQVCIAWSS SSCHDGKAWL HVCITGDDKN ATASFIYNGR LVDSVVSWSN DILRTQESEC VCINGTCTVV MT DGNATGK ADTKILFIEE GKIVHTSKLS GSAQHVEECS CYPRYPGVRC VCRDNWKGSN RPIIDINIKD HSIVSRYVCS GLV GDTPRK SDSSSSSHCL NPNNEKGDHG VKGWAFDDGN DVWMGRTINE TSRLGYETFK VVEGWSNPKS KLQINRQVIV DRGD RSGYS GIFSVEGKSC INRCFYVELI RGRKEETEVL WTSNSIVVFC GTSGTYGTGS WPDGANLSLM HI UniProtKB: Neuraminidase |
-Macromolecule #2: Fab NDS.3, heavy chain
| Macromolecule | Name: Fab NDS.3, heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.20924 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKPGGSLRL SCAASGFTFS DYSMTWIRQI PGKGLEWLSY TTSSGRPIFY ADSVKGRFTM SRDNAKMSVY LQMNSLRDA DSAVYYCARG DPHYIWGTYL DSWGPGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL ...String: EVQLVESGGG LVKPGGSLRL SCAASGFTFS DYSMTWIRQI PGKGLEWLSY TTSSGRPIFY ADSVKGRFTM SRDNAKMSVY LQMNSLRDA DSAVYYCARG DPHYIWGTYL DSWGPGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDK |
-Macromolecule #3: Fab NDS.3, light chain
| Macromolecule | Name: Fab NDS.3, light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.863139 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPPSA SGSPGQSVTI SCTGTTSDFG DHNYVSWYQQ RPGEAPKLII YDVSKRPSGV PDRFSGSKSG NTASLTVSRL QADDEANYY CSSIEGSNTL LFGGGTKLTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: QSALTQPPSA SGSPGQSVTI SCTGTTSDFG DHNYVSWYQQ RPGEAPKLII YDVSKRPSGV PDRFSGSKSG NTASLTVSRL QADDEANYY CSSIEGSNTL LFGGGTKLTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: SwissModel / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
| Output model |  PDB-8gav: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)