[English] 日本語
 Yorodumi
Yorodumi- EMDB-29886: Partial DNA termination subcomplex of Xenopus laevis DNA polymera... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | ||||||||||||
 Map data Map data | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species | |||||||||||||
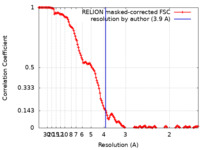
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Mullins EA / Chazin WJ / Eichman BF | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: A mechanistic model of primer synthesis from catalytic structures of DNA polymerase α-primase. Authors: Elwood A Mullins / Lauren E Salay / Clarissa L Durie / Noah P Bradley / Jane E Jackman / Melanie D Ohi / Walter J Chazin / Brandt F Eichman Abstract: The mechanism by which polymerase α-primase (polα-primase) synthesizes chimeric RNA-DNA primers of defined length and composition, necessary for replication fidelity and genome stability, is ...The mechanism by which polymerase α-primase (polα-primase) synthesizes chimeric RNA-DNA primers of defined length and composition, necessary for replication fidelity and genome stability, is unknown. Here, we report cryo-EM structures of polα-primase in complex with primed templates representing various stages of DNA synthesis. Our data show how interaction of the primase regulatory subunit with the primer 5'-end facilitates handoff of the primer to polα and increases polα processivity, thereby regulating both RNA and DNA composition. The structures detail how flexibility within the heterotetramer enables synthesis across two active sites and provide evidence that termination of DNA synthesis is facilitated by reduction of polα and primase affinities for the varied conformations along the chimeric primer/template duplex. Together, these findings elucidate a critical catalytic step in replication initiation and provide a comprehensive model for primer synthesis by polα-primase. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29886.map.gz emd_29886.map.gz | 121.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29886-v30.xml emd-29886-v30.xml emd-29886.xml emd-29886.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29886_fsc.xml emd_29886_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_29886.png emd_29886.png | 19 KB | ||
| Others |  emd_29886_half_map_1.map.gz emd_29886_half_map_1.map.gz emd_29886_half_map_2.map.gz emd_29886_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29886 http://ftp.pdbj.org/pub/emdb/structures/EMD-29886 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29886 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29886 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29886.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29886.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Partial DNA termination subcomplex of Xenopus laevis DNA...
| File | emd_29886_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase (half 2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
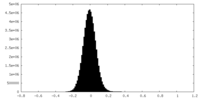
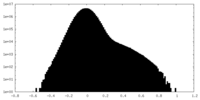
| Density Histograms |
-Half map: Partial DNA termination subcomplex of Xenopus laevis DNA...
| File | emd_29886_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase (half 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
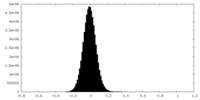
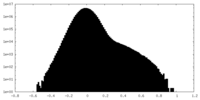
| Density Histograms |
- Sample components
Sample components
-Entire : Polymerase alpha-primase with a DNA termination substrate
| Entire | Name: Polymerase alpha-primase with a DNA termination substrate |
|---|---|
| Components |
|
-Supramolecule #1: Polymerase alpha-primase with a DNA termination substrate
| Supramolecule | Name: Polymerase alpha-primase with a DNA termination substrate type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all Details: Heterotetrameric protein complex with an RNA-DNA/DNA duplex |
|---|---|
| Molecular weight | Theoretical: 330 KDa |
-Supramolecule #2: Polymerase alpha
| Supramolecule | Name: Polymerase alpha / type: complex / ID: 2 / Chimera: Yes / Parent: 1 / Macromolecule list: #1 / Details: Heterodimer consisting of POLA1 and POLA2 |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 200 KDa |
-Supramolecule #3: Primase
| Supramolecule | Name: Primase / type: complex / ID: 3 / Chimera: Yes / Parent: 1 / Details: Heterodimer consisting of PRIM1 and PRIM2 |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 110 KDa |
-Supramolecule #4: DNA termination substrate
| Supramolecule | Name: DNA termination substrate / type: complex / ID: 4 / Chimera: Yes / Parent: 1 / Macromolecule list: #2-#3 / Details: RNA-DNA/DNA duplex |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 30 KDa |
-Macromolecule #1: POLA1
| Macromolecule | Name: POLA1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: SNAADGSQVF RFYWLDAYED QYSQPGVVYL FGKVWIESAD AYVSCCVSVK NIERTVYLLP RENRVQLSTG KDTGAPVSMM HVYQEFNEAV AEKYKIMKFK SKKVDKDYAF EIPDVPASSE YLEVRYSADS PQLPQDLKGE TFSHVFGTNT SSLELFLLSR KIKGPSWLEI ...String: SNAADGSQVF RFYWLDAYED QYSQPGVVYL FGKVWIESAD AYVSCCVSVK NIERTVYLLP RENRVQLSTG KDTGAPVSMM HVYQEFNEAV AEKYKIMKFK SKKVDKDYAF EIPDVPASSE YLEVRYSADS PQLPQDLKGE TFSHVFGTNT SSLELFLLSR KIKGPSWLEI KSPQLSSQPM SWCKVEAVVT RPDQVSVVKD LAPPPVVVLS LSMKTVQNAK THQNEIVAIA ALVHHTFPLD KAPPQPPFQT HFCVLSKLND CIFPYDYNEA VKQKNANIEI ALTERTLLGF FLAKIHKIDP DVIVGHDIYG FDLEVLLQRI NSCKVPFWSK IGRLRRSVMP KLGGRSGFAE RNAACGRIIC DIEISAKELI RCKSYHLSEL VHQILKAERV VIPPENIRNA YNDSVHLLYM LENTWIDAKF ILQIMCELNV LPLALQITNI AGNVMSRTLM GGRSERNEYL LLHAFTENNF IVPDKPVFKK MQQTTVEDND DMGTDQNKNK SRKKAAYAGG LVLEPKVGFY DKFILLLDFN SLYPSIIQEY NICFTTVHRE APSTQKGEDQ DEIPELPHSD LEMGILPREI RKLVERRRHV KQLMKQPDLN PDLYLQYDIR QKALKLTANS MYGCLGFSYS RFYAKPLAAL VTHQGREILL HTKEMVQKMN LEVIYGDTDS IMINTNCNNL EEVFKLGNRV KSEINKSYKL LEIDIDGIFK SLLLLKKKKY AALTVEPTGD GKYVTKQELK GLDIVRRDWC ELAKQAGNYV ISQILSDQPR DSIVENIQKK LTEIGENVTN GTVPITQYEI NKALTKDPQD YPDKKSLPHV HVALWINSQG GRKVKAGDTI SYVICQDGSN LSASQRAYAQ EQLQKQENLS IDTQYYLSQQ VHPVVARICE PIDGIDSALI AMWLGLDPSQ FRAHRHYQQD EENDALLGGP SQLTDEEKYR DCERFKFFCP KCGTENIYDN VFDGSGLQIE PGLKRCSKPE CDASPLDYVI QVHNKLLLDI RRYIKKYYSG WLVCEEKTCQ NRTRRLPLSF SRNGPICQAC SKATLRSEYP EKALYTQLCF YRFIFDWDYA LEKVVSEQER GHLKKKLFQE SENQYKKLKS TVDQVLSRSG YSEVNLSKLF QTLNTIK |
-Macromolecule #2: DNA template
| Macromolecule | Name: DNA template / type: dna / ID: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: (DT)(DG)(DT)(DA)(DT)(DG)(DT)(DA)(DT)(DG) (DT)(DA)(DT)(DG)(DT)(DC)(DG)(DC)(DT)(DA) (DC)(DA)(DA)(DT)(DC)(DG)(DC)(DT)(DA)(DA) (DG)(DT)(DT)(DC)(DA)(DC)(DG)(DC)(DA)(DG) (DT)(DA)(DT)(DC)(DC)(DT)(DG) ...String: (DT)(DG)(DT)(DA)(DT)(DG)(DT)(DA)(DT)(DG) (DT)(DA)(DT)(DG)(DT)(DC)(DG)(DC)(DT)(DA) (DC)(DA)(DA)(DT)(DC)(DG)(DC)(DT)(DA)(DA) (DG)(DT)(DT)(DC)(DA)(DC)(DG)(DC)(DA)(DG) (DT)(DA)(DT)(DC)(DC)(DT)(DG)(DT)(DA)(DT) (DG)(DT)(DA)(DT)(DG)(DT)(DA)(DT)(DG) |
-Macromolecule #3: RNA-DNA primer
| Macromolecule | Name: RNA-DNA primer / type: dna / ID: 3 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: (GTP)GAUACUGC(DG) (DT)(DG)(DA)(DA)(DC)(DT)(DT)(DA)(DG)(DC) (DG)(DA)(DT)(DT)(DG)(DT)(DA)(DG)(DOC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 22893 / Average electron dose: 55.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)