[English] 日本語
 Yorodumi
Yorodumi- EMDB-29857: Molecular mechanism of nucleotide inhibition of human uncoupling ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | SLC25 / mitochondrial carrier / uncoupling / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpurine ribonucleotide binding / cellular response to dehydroepiandrosterone / Mitochondrial Uncoupling / The fatty acid cycling model / oxidative phosphorylation uncoupler activity / mitochondrial transmembrane transport / adaptive thermogenesis / cardiolipin binding / cellular response to fatty acid / regulation of reactive oxygen species biosynthetic process ...purine ribonucleotide binding / cellular response to dehydroepiandrosterone / Mitochondrial Uncoupling / The fatty acid cycling model / oxidative phosphorylation uncoupler activity / mitochondrial transmembrane transport / adaptive thermogenesis / cardiolipin binding / cellular response to fatty acid / regulation of reactive oxygen species biosynthetic process / response to temperature stimulus / long-chain fatty acid binding / cellular response to cold / diet induced thermogenesis / detection of maltose stimulus / maltose transport complex / proton transmembrane transporter activity / carbohydrate transport / transmembrane transporter activity / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / brown fat cell differentiation / Transcriptional regulation of brown and beige adipocyte differentiation by EBF2 / cellular response to hormone stimulus / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / proton transmembrane transport / ATP-binding cassette (ABC) transporter complex / response to cold / cell chemotaxis / cellular response to reactive oxygen species / response to nutrient levels / GDP binding / positive regulation of cold-induced thermogenesis / outer membrane-bounded periplasmic space / periplasmic space / mitochondrial inner membrane / DNA damage response / regulation of transcription by RNA polymerase II / GTP binding / mitochondrion / membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||||||||
 Authors Authors | Gogoi P / Jones SA / Ruprecht JJ / King MS / Lee Y / Zogg T / Pardon E / Chand D / Steimle S / Copeman D ...Gogoi P / Jones SA / Ruprecht JJ / King MS / Lee Y / Zogg T / Pardon E / Chand D / Steimle S / Copeman D / Cotrim CA / Steyaert J / Crichton PG / Moiseenkova-Bell V / Kunji ERS | |||||||||||||||||||||
| Funding support |  United States, United States,  United Kingdom, European Union, United Kingdom, European Union,  Belgium, 6 items Belgium, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural basis of purine nucleotide inhibition of human uncoupling protein 1. Authors: Scott A Jones / Prerana Gogoi / Jonathan J Ruprecht / Martin S King / Yang Lee / Thomas Zögg / Els Pardon / Deepak Chand / Stefan Steimle / Danielle M Copeman / Camila A Cotrim / Jan ...Authors: Scott A Jones / Prerana Gogoi / Jonathan J Ruprecht / Martin S King / Yang Lee / Thomas Zögg / Els Pardon / Deepak Chand / Stefan Steimle / Danielle M Copeman / Camila A Cotrim / Jan Steyaert / Paul G Crichton / Vera Moiseenkova-Bell / Edmund R S Kunji /    Abstract: Mitochondrial uncoupling protein 1 (UCP1) gives brown adipose tissue of mammals its specialized ability to burn calories as heat for thermoregulation. When activated by fatty acids, UCP1 catalyzes ...Mitochondrial uncoupling protein 1 (UCP1) gives brown adipose tissue of mammals its specialized ability to burn calories as heat for thermoregulation. When activated by fatty acids, UCP1 catalyzes the leak of protons across the mitochondrial inner membrane, short-circuiting the mitochondrion to generate heat, bypassing ATP synthesis. In contrast, purine nucleotides bind and inhibit UCP1, regulating proton leak by a molecular mechanism that is unclear. We present the cryo-electron microscopy structure of the GTP-inhibited state of UCP1, which is consistent with its nonconducting state. The purine nucleotide cross-links the transmembrane helices of UCP1 with an extensive interaction network. Our results provide a structural basis for understanding the specificity and pH dependency of the regulatory mechanism. UCP1 has retained all of the key functional and structural features required for a mitochondrial carrier-like transport mechanism. The analysis shows that inhibitor binding prevents the conformational changes that UCP1 uses to facilitate proton leak. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29857.map.gz emd_29857.map.gz | 398.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29857-v30.xml emd-29857-v30.xml emd-29857.xml emd-29857.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29857.png emd_29857.png | 27.4 KB | ||
| Filedesc metadata |  emd-29857.cif.gz emd-29857.cif.gz | 6.9 KB | ||
| Others |  emd_29857_half_map_1.map.gz emd_29857_half_map_1.map.gz emd_29857_half_map_2.map.gz emd_29857_half_map_2.map.gz | 391.9 MB 391.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29857 http://ftp.pdbj.org/pub/emdb/structures/EMD-29857 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29857 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29857 | HTTPS FTP |
-Validation report
| Summary document |  emd_29857_validation.pdf.gz emd_29857_validation.pdf.gz | 885.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29857_full_validation.pdf.gz emd_29857_full_validation.pdf.gz | 884.7 KB | Display | |
| Data in XML |  emd_29857_validation.xml.gz emd_29857_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  emd_29857_validation.cif.gz emd_29857_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29857 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29857 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29857 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29857 | HTTPS FTP |
-Related structure data
| Related structure data |  8g8wMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29857.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29857.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29857_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
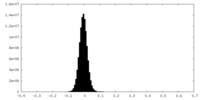
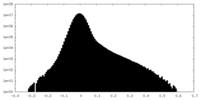
| Density Histograms |
-Half map: #1
| File | emd_29857_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
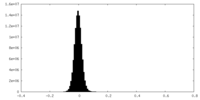
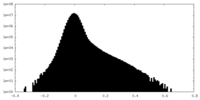
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of human uncoupling protein 1 with two promacrobodies
| Entire | Name: Complex of human uncoupling protein 1 with two promacrobodies |
|---|---|
| Components |
|
-Supramolecule #1: Complex of human uncoupling protein 1 with two promacrobodies
| Supramolecule | Name: Complex of human uncoupling protein 1 with two promacrobodies type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Mitochondrial brown fat uncoupling protein 1
| Macromolecule | Name: Mitochondrial brown fat uncoupling protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.339629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TSEDGGLTAS DVHPTLGVQL FSAGIAACLA DVITFPLDTA KVRLQVQGEC PTSSVIRYKG VLGTITAVVK TEGRMKLYSG LPAGLQRQI SSASLRIGLY DTVQEFLTAG KETAPSLGSK ILAGLTTGGV AVFIGQPTEV VKVRLQAQSH LHGIKPRYTG T YNAYRIIA ...String: TSEDGGLTAS DVHPTLGVQL FSAGIAACLA DVITFPLDTA KVRLQVQGEC PTSSVIRYKG VLGTITAVVK TEGRMKLYSG LPAGLQRQI SSASLRIGLY DTVQEFLTAG KETAPSLGSK ILAGLTTGGV AVFIGQPTEV VKVRLQAQSH LHGIKPRYTG T YNAYRIIA TTEGLTGLWK GTTPNLMRSV IINCTELVTY DLMKEAFVKN NILADDVPCH LVSALIAGFC ATAMSSPVDV VK TRFINSP PGQYKSVPNC AMKVFTNEGP TAFFKGLVPS FLRLGSWNVI MFVCFEQLKR ELSKSRQTMD CAT UniProtKB: Mitochondrial brown fat uncoupling protein 1 |
-Macromolecule #2: Pro-macrobody 71, Maltose/maltodextrin-binding periplasmic protei...
| Macromolecule | Name: Pro-macrobody 71, Maltose/maltodextrin-binding periplasmic protein chimera type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.45391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPSQVQLVES GGGLVQAGDS LRLSCAASGL TLKNYAMGWF RQAPGKEHEF VAVISWSGSG TSYADSVEGR FTISRDNAKN TAFLQMSSL KPEDTAVYYC AARDGGYGSR WPDEYTYWGQ GTQVTVPPLV IWINGDKGYN GLAEVGKKFE KDTGIKVTVE H PDKLEEKF ...String: GPSQVQLVES GGGLVQAGDS LRLSCAASGL TLKNYAMGWF RQAPGKEHEF VAVISWSGSG TSYADSVEGR FTISRDNAKN TAFLQMSSL KPEDTAVYYC AARDGGYGSR WPDEYTYWGQ GTQVTVPPLV IWINGDKGYN GLAEVGKKFE KDTGIKVTVE H PDKLEEKF PQVAATGDGP DIIFWAHDRF GGYAQSGLLA EITPDKAFQD KLYPFTWDAV RYNGKLIAYP IAVEALSLIY NK DLLPNPP KTWEEIPALD KELKAKGKSA LMFNLQEPYF TWPLIAADGG YAFKYENGKY DIKDVGVDNA GAKAGLTFLV DLI KNKHMN ADTDYSIAEA AFNKGETAMT INGPWAWSNI DTSKVNYGVT VLPTFKGQPS KPFVGVLSAG INAASPNKEL AKEF LENYL LTDEGLEAVN KDKPLGAVAL KSYEEELAKD PRIAATMENA QKGEIMPNIP QMSAFWYAVR TAVINAASGR QTVDE ALKD AQTPGS UniProtKB: Maltose/maltodextrin-binding periplasmic protein |
-Macromolecule #3: Pro-Macrobody 65, Maltose/maltodextrin-binding periplasmic protei...
| Macromolecule | Name: Pro-Macrobody 65, Maltose/maltodextrin-binding periplasmic protein chimera type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.05282 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPSQVQLVES GGGLVQAGGS LRLSCAPSGR TSSTYTMGWF RQAPGKEREF VAAISWTGTP YYADSVKGRF TISRDNAKNT VYLQMNSLK PEDTAVYYCA AARPGLFIFV SDYARTAKYD YWGQGTQVTV PPLVIWINGD KGYNGLAEVG KKFEKDTGIK V TVEHPDKL ...String: GPSQVQLVES GGGLVQAGGS LRLSCAPSGR TSSTYTMGWF RQAPGKEREF VAAISWTGTP YYADSVKGRF TISRDNAKNT VYLQMNSLK PEDTAVYYCA AARPGLFIFV SDYARTAKYD YWGQGTQVTV PPLVIWINGD KGYNGLAEVG KKFEKDTGIK V TVEHPDKL EEKFPQVAAT GDGPDIIFWA HDRFGGYAQS GLLAEITPDK AFQDKLYPFT WDAVRYNGKL IAYPIAVEAL SL IYNKDLL PNPPKTWEEI PALDKELKAK GKSALMFNLQ EPYFTWPLIA ADGGYAFKYE NGKYDIKDVG VDNAGAKAGL TFL VDLIKN KHMNADTDYS IAEAAFNKGE TAMTINGPWA WSNIDTSKVN YGVTVLPTFK GQPSKPFVGV LSAGINAASP NKEL AKEFL ENYLLTDEGL EAVNKDKPLG AVALKSYEEE LAKDPRIAAT MENAQKGEIM PNIPQMSAFW YAVRTAVINA ASGRQ TVDE ALKDAQTPGS UniProtKB: Maltose/maltodextrin-binding periplasmic protein |
-Macromolecule #5: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #6: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 6 / Number of copies: 3 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.14 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 72.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 203799 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)