[English] 日本語
 Yorodumi
Yorodumi- EMDB-29834: Time-resolved cryo-EM study of the 70S recycling by the HflX:1st ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 50S focused from 30S subtracted | |||||||||
 Map data Map data | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 50S focused from 30S subtracted | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Recycling / Time-resolved Cryo-EM / 70S / HflX / RIBOSOME | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Bhattacharjee S / Brown PZ / Frank J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling. Authors: Sayan Bhattacharjee / Xiangsong Feng / Suvrajit Maji / Prikshat Dadhwal / Zhening Zhang / Zuben P Brown / Joachim Frank /  Abstract: The rapid kinetics of biological processes and associated short-lived conformational changes pose a significant challenge in attempts to structurally visualize biomolecules during a reaction in real ...The rapid kinetics of biological processes and associated short-lived conformational changes pose a significant challenge in attempts to structurally visualize biomolecules during a reaction in real time. Conventionally, on-pathway intermediates have been trapped using chemical modifications or reduced temperature, giving limited insights. Here, we introduce a time-resolved cryo-EM method using a reusable PDMS-based microfluidic chip assembly with high reactant mixing efficiency. Coating of PDMS walls with SiO virtually eliminates non-specific sample adsorption and ensures maintenance of the stoichiometry of the reaction, rendering it highly reproducible. In an operating range from 10 to 1,000 ms, the device allows us to follow in vitro reactions of biological molecules at resolution levels in the range of 3 Å. By employing this method, we show the mechanism of progressive HflX-mediated splitting of the 70S E. coli ribosome in the presence of the GTP via capture of three high-resolution reaction intermediates within 140 ms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29834.map.gz emd_29834.map.gz | 141.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29834-v30.xml emd-29834-v30.xml emd-29834.xml emd-29834.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29834_fsc.xml emd_29834_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_29834.png emd_29834.png | 99.4 KB | ||
| Masks |  emd_29834_msk_1.map emd_29834_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29834.cif.gz emd-29834.cif.gz | 3.9 KB | ||
| Others |  emd_29834_half_map_1.map.gz emd_29834_half_map_1.map.gz emd_29834_half_map_2.map.gz emd_29834_half_map_2.map.gz | 139.3 MB 139.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29834 http://ftp.pdbj.org/pub/emdb/structures/EMD-29834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29834 | HTTPS FTP |
-Validation report
| Summary document |  emd_29834_validation.pdf.gz emd_29834_validation.pdf.gz | 1.4 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29834_full_validation.pdf.gz emd_29834_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  emd_29834_validation.xml.gz emd_29834_validation.xml.gz | 19.6 KB | Display | |
| Data in CIF |  emd_29834_validation.cif.gz emd_29834_validation.cif.gz | 25.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29834 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29834 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29834 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29834 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29834.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29834.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 50S focused from 30S subtracted | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02529 Å | ||||||||||||||||||||||||||||||||||||
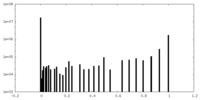
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29834_msk_1.map emd_29834_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
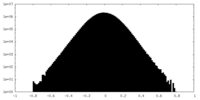
| Density Histograms |
-Half map: Time-resolved cryo-EM study of the 70S recycling by...
| File | emd_29834_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 50S focused from 30S subtracted, half-I | ||||||||||||
| Projections & Slices |
| ||||||||||||
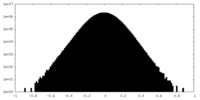
| Density Histograms |
-Half map: Time-resolved cryo-EM study of the 70S recycling by...
| File | emd_29834_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 50S focused from 30S subtracted, half-II | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Apo 70S 1st intermediate
| Entire | Name: Apo 70S 1st intermediate |
|---|---|
| Components |
|
-Supramolecule #1: Apo 70S 1st intermediate
| Supramolecule | Name: Apo 70S 1st intermediate / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)