[English] 日本語
 Yorodumi
Yorodumi- EMDB-29741: Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ligand-gated ion channel / cys-loop receptor / neurotransmitter receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationGABA receptor activation / inhibitory synapse / cellular response to histamine / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation / postsynaptic specialization membrane ...GABA receptor activation / inhibitory synapse / cellular response to histamine / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation / postsynaptic specialization membrane / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / chloride channel activity / adult behavior / cochlea development / chloride channel complex / neuron development / cytoplasmic vesicle membrane / chloride transmembrane transport / post-embryonic development / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / GABA-ergic synapse / cytoplasmic vesicle / chemical synaptic transmission / dendritic spine / axon / dendrite / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
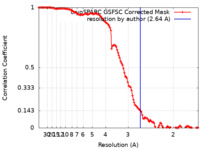
| Method | single particle reconstruction / cryo EM / Resolution: 2.64 Å | |||||||||
 Authors Authors | Sun C / Gouaux E | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation | Journal: Acta Crystallogr D Struct Biol / Year: 2018 Title: Real-space refinement in PHENIX for cryo-EM and crystallography. Authors: Pavel V Afonine / Billy K Poon / Randy J Read / Oleg V Sobolev / Thomas C Terwilliger / Alexandre Urzhumtsev / Paul D Adams /    Abstract: This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast ...This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast calculation, which in turn makes it possible to identify optimal data-restraint weights as part of routine refinements with little runtime cost. Refinement of atomic models against low-resolution data benefits from the inclusion of as much additional information as is available. In addition to standard restraints on covalent geometry, phenix.real_space_refine makes use of extra information such as secondary-structure and rotamer-specific restraints, as well as restraints or constraints on internal molecular symmetry. The re-refinement of 385 cryo-EM-derived models available in the Protein Data Bank at resolutions of 6 Å or better shows significant improvement of the models and of the fit of these models to the target maps. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29741.map.gz emd_29741.map.gz | 86 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29741-v30.xml emd-29741-v30.xml emd-29741.xml emd-29741.xml | 30.2 KB 30.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29741_fsc.xml emd_29741_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29741.png emd_29741.png | 178 KB | ||
| Masks |  emd_29741_msk_1.map emd_29741_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29741.cif.gz emd-29741.cif.gz | 9.4 KB | ||
| Others |  emd_29741_half_map_1.map.gz emd_29741_half_map_1.map.gz emd_29741_half_map_2.map.gz emd_29741_half_map_2.map.gz | 165.3 MB 165.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29741 http://ftp.pdbj.org/pub/emdb/structures/EMD-29741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29741 | HTTPS FTP |
-Validation report
| Summary document |  emd_29741_validation.pdf.gz emd_29741_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29741_full_validation.pdf.gz emd_29741_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_29741_validation.xml.gz emd_29741_validation.xml.gz | 20.2 KB | Display | |
| Data in CIF |  emd_29741_validation.cif.gz emd_29741_validation.cif.gz | 25.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29741 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29741 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29741 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29741 | HTTPS FTP |
-Related structure data
| Related structure data |  8g5fMC  8foiC  8g4nC  8g4oC  8g4xC  8g5gC  8g5hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29741.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29741.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
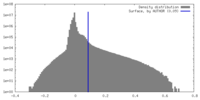
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29741_msk_1.map emd_29741_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29741_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
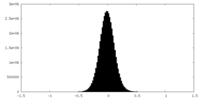
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29741_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
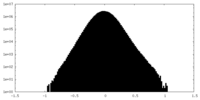
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Native GABAA Receptor isolated from mouse brain containing one al...
+Supramolecule #1: Native GABAA Receptor isolated from mouse brain containing one al...
+Macromolecule #1: Gamma-aminobutyric acid receptor subunit alpha-3
+Macromolecule #2: Gamma-aminobutyric acid receptor subunit beta-2
+Macromolecule #3: Gamma-aminobutyric acid receptor subunit alpha-1
+Macromolecule #4: Gamma-aminobutyric acid receptor subunit gamma-2
+Macromolecule #5: Heavy Chain of 8E3 Fab
+Macromolecule #6: Light Chain of 8E3 Fab
+Macromolecule #10: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(o...
+Macromolecule #11: allopregnanolone
+Macromolecule #12: GAMMA-AMINO-BUTANOIC ACID
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8g5f: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)