[English] 日本語
 Yorodumi
Yorodumi- EMDB-29732: Cryo-EM consensus structure of Escherichia coli que-PEC (paused e... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | RNA Polymerase / Riboswitch / Transcription / preQ1 | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-directed RNA polymerase complex / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
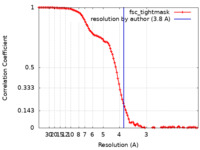
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||
 Authors Authors | Porta JC / Chauvier A / Deb I / Ellinger E / Frank AT / Meze K / Ohi MD / Walter NG | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand. Authors: Adrien Chauvier / Jason C Porta / Indrajit Deb / Emily Ellinger / Katarina Meze / Aaron T Frank / Melanie D Ohi / Nils G Walter /   Abstract: Folding of nascent transcripts can be modulated by the RNA polymerase (RNAP) that carries out their transcription, and vice versa. A pause of RNAP during transcription of a preQ riboswitch (termed ...Folding of nascent transcripts can be modulated by the RNA polymerase (RNAP) that carries out their transcription, and vice versa. A pause of RNAP during transcription of a preQ riboswitch (termed que-PEC) is stabilized by a previously characterized template consensus sequence and the ligand-free conformation of the nascent RNA. Ligand binding to the riboswitch induces RNAP pause release and downstream transcription termination; however, the mechanism by which riboswitch folding modulates pausing is unclear. Here, we report single-particle cryo-electron microscopy reconstructions of que-PEC in ligand-free and ligand-bound states. In the absence of preQ, the RNA transcript is in an unexpected hyper-translocated state, preventing downstream nucleotide incorporation. Strikingly, on ligand binding, the riboswitch rotates around its helical axis, expanding the surrounding RNAP exit channel and repositioning the transcript for elongation. Our study reveals the tight coupling by which nascent RNA structures and their ligands can functionally regulate the macromolecular transcription machinery. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29732.map.gz emd_29732.map.gz | 95.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29732-v30.xml emd-29732-v30.xml emd-29732.xml emd-29732.xml | 29 KB 29 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29732_fsc.xml emd_29732_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29732.png emd_29732.png | 52.6 KB | ||
| Filedesc metadata |  emd-29732.cif.gz emd-29732.cif.gz | 8.9 KB | ||
| Others |  emd_29732_half_map_1.map.gz emd_29732_half_map_1.map.gz emd_29732_half_map_2.map.gz emd_29732_half_map_2.map.gz | 93.4 MB 93.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29732 http://ftp.pdbj.org/pub/emdb/structures/EMD-29732 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29732 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29732 | HTTPS FTP |
-Related structure data
| Related structure data |  8g4wMC  8f3cC  8g00C  8g1sC  8g2wC  8g7eC  8g8zC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29732.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29732.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29732_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29732_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Cryo-EM consensus structure of Escherichia coli que-PEC (paused e...
+Supramolecule #1: Cryo-EM consensus structure of Escherichia coli que-PEC (paused e...
+Macromolecule #1: DNA (39-mer)
+Macromolecule #2: DNA (31-MER)
+Macromolecule #3: DNA-directed RNA polymerase subunit alpha
+Macromolecule #4: DNA-directed RNA polymerase subunit omega
+Macromolecule #6: DNA-directed RNA polymerase subunit beta
+Macromolecule #7: DNA-directed RNA polymerase subunit beta'
+Macromolecule #5: RNA (47-MER)
+Macromolecule #8: 7-DEAZA-7-AMINOMETHYL-GUANINE
+Macromolecule #9: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.00 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE Details: VITRIFICATION WAS CARRIED OUT IN A CHAMBER WITH THE TEMPERATURE SET TO 4 DEGREES CELSIUS.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Dimensions - Width: 1 pixel / Digitization - Dimensions - Height: 1 pixel / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-8g4w: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)