Yorodumi
Yorodumi+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 48-nm doublet microtubule from Tetrahymena thermophila strain CU428 | |||||||||||||||
 Map data Map data | composite map of 48-nm doublet from Tetrahymena strain CU428 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Cilia / Axoneme / Doublet Microtubule / Microtubule Inner Protein / STRUCTURAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcilium-dependent cell motility / axoneme assembly / axonemal microtubule / nucleoside-diphosphate kinase / UTP biosynthetic process / CTP biosynthetic process / nucleoside diphosphate kinase activity / GTP biosynthetic process / motile cilium / mitotic cytokinesis ...cilium-dependent cell motility / axoneme assembly / axonemal microtubule / nucleoside-diphosphate kinase / UTP biosynthetic process / CTP biosynthetic process / nucleoside diphosphate kinase activity / GTP biosynthetic process / motile cilium / mitotic cytokinesis / cilium assembly / axoneme / alpha-tubulin binding / microtubule-based process / Hsp70 protein binding / cell projection / mitotic spindle organization / meiotic cell cycle / Hsp90 protein binding / structural constituent of cytoskeleton / mitotic spindle / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / cytoskeleton / calmodulin binding / cilium / hydrolase activity / GTPase activity / calcium ion binding / GTP binding / zinc ion binding / ATP binding / metal ion binding / nucleus / cytoplasm Similarity search - Function CCDC81, HU domain 1 / CCDC81, HU domain 2 / CCDC81 eukaryotic HU domain 1 / CCDC81 eukaryotic HU domain 2 / Sperm-tail PG-rich repeat / Sperm-tail PG-rich repeat / : / : / : / : ...CCDC81, HU domain 1 / CCDC81, HU domain 2 / CCDC81 eukaryotic HU domain 1 / CCDC81 eukaryotic HU domain 2 / Sperm-tail PG-rich repeat / Sperm-tail PG-rich repeat / : / : / : / : / Uncharacterised protein FAM166/UPF0605 / Cilia- and flagella-associated protein 77 / Ciliary microtubule inner protein 2B-like / Cilia- and flagella-associated protein 77 / Nucleoside diphosphate kinase 7 / Piercer of microtubule wall 1/2 / Piercer of microtubule wall 1/2 / Cilia- and flagella-associated protein 141 / Cilia- and flagella-associated protein 141 / Cilia- and flagella-associated protein 161-like domain / : / EFHB C-terminal EF-hand domain / : / Nucleoside diphosphate kinase 7 N-terminal PH domain / Meiosis-specific nuclear structural protein 1 / Cilia- and flagella-associated protein 45 / NDPK7, second NDPk domain / Cilia- and flagella- associated protein 210 / CFAP53/TCHP / Trichohyalin-plectin-homology domain / Trichohyalin-plectin-homology domain / Dienelactone hydrolase / RIB43A / RIB43A / DM10 domain / EF-hand domain-containing protein EFHC1/EFHC2/EFHB / DM10 domain / DM10 domain profile. / Domains in hypothetical proteins in Drosophila, C. elegans and mammals. Occurs singly in some nucleoside diphosphate kinases. / Enkurin domain / : / Calmodulin-binding / Enkurin domain profile. / : / CFA20 domain / Cilia- and flagella-associated protein 20/CFAP20DC / CFA20 domain / Dienelactone hydrolase family / Parkin co-regulated protein / Parkin co-regulated protein / Nucleoside diphosphate kinase, active site / Nucleoside diphosphate kinase (NDPK) active site signature. / Nucleoside diphosphate kinase / Nucleoside diphosphate kinase (NDPK)-like domain profile. / Nucleoside diphosphate kinase-like domain / Nucleoside diphosphate kinase / NDK / Nucleoside diphosphate kinase-like domain superfamily / EF-hand domain pair / Mir domain superfamily / Alpha tubulin / Tubulin-beta mRNA autoregulation signal. / Beta tubulin, autoregulation binding site / Beta tubulin / Tubulin / Tubulin, C-terminal / Tubulin C-terminal domain / Tubulin, conserved site / Tubulin subunits alpha, beta, and gamma signature. / Tubulin/FtsZ family, C-terminal domain / Tubulin/FtsZ-like, C-terminal domain / Tubulin/FtsZ, C-terminal / Tubulin/FtsZ, 2-layer sandwich domain / Tubulin/FtsZ family, GTPase domain / Tubulin/FtsZ family, GTPase domain / Tubulin/FtsZ, GTPase domain / Tubulin/FtsZ, GTPase domain superfamily / EF-hand domain pair / EF-hand, calcium binding motif / EF-Hand 1, calcium-binding site / EF-hand calcium-binding domain. / EF-hand calcium-binding domain profile. / EF-hand domain / EF-hand domain pair / Alpha/Beta hydrolase fold / WD domain, G-beta repeat / Armadillo-type fold / WD40 repeat, conserved site / Trp-Asp (WD) repeats signature. / Trp-Asp (WD) repeats profile. / Trp-Asp (WD) repeats circular profile. / WD40 repeats / WD40 repeat / WD40-repeat-containing domain superfamily / WD40/YVTN repeat-like-containing domain superfamily Similarity search - Domain/homology Flagellar FliJ protein / Uncharacterized protein / RIB43A protein / EF hand protein / Uncharacterized protein / Meiosis-specific nuclear structural protein 1 / Flagellar microtugule protofilament ribbon protein / DNA polymerase delta C4-type zinc-finger protein / Sperm-tail PG-rich repeat protein / Parkin co-regulated protein ...Flagellar FliJ protein / Uncharacterized protein / RIB43A protein / EF hand protein / Uncharacterized protein / Meiosis-specific nuclear structural protein 1 / Flagellar microtugule protofilament ribbon protein / DNA polymerase delta C4-type zinc-finger protein / Sperm-tail PG-rich repeat protein / Parkin co-regulated protein / Ptype ATPase, putative / Uncharacterized protein / EF-hand protein / EF-hand pair protein / EF-hand pair protein / EFHB C-terminal EF-hand domain-containing protein / Parkin co-regulated protein / Uncharacterized protein / Tubulin alpha chain / Tubulin beta chain / Uncharacterized protein / Uncharacterized protein / Transcription factor iib protein, putative / EF hand protein, putative / Sperm-tail PG-rich repeat protein / Cilia- and flagella-associated protein 52 / Uncharacterized protein / Uncharacterized protein / Dienelactone hydrolase family protein / Ciliary microtubule inner protein 2A-C-like domain-containing protein / Uncharacterized protein / Uncharacterized protein / Ciliary microtubule inner protein 2A-C-like domain-containing protein / Trichohyalin-plectin-homology domain-containing protein / STOP protein / EF hand protein / Cilia- and flagella-associated protein 53 / Protofilament ribbon protein / Uncharacterized protein / Sperm-tail PG-rich repeat protein / Cilia- and flagella-associated protein 45 / Nucleoside diphosphate kinase Similarity search - Component | |||||||||||||||
| Biological species |  | |||||||||||||||
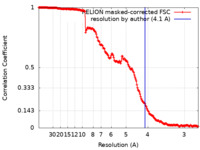
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||||||||
 Authors Authors | Black CS / Kubo S / Yang SK / Bui KH | |||||||||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins. Authors: Shintaroh Kubo / Corbin S Black / Ewa Joachimiak / Shun Kai Yang / Thibault Legal / Katya Peri / Ahmad Abdelzaher Zaki Khalifa / Avrin Ghanaeian / Caitlyn L McCafferty / Melissa Valente- ...Authors: Shintaroh Kubo / Corbin S Black / Ewa Joachimiak / Shun Kai Yang / Thibault Legal / Katya Peri / Ahmad Abdelzaher Zaki Khalifa / Avrin Ghanaeian / Caitlyn L McCafferty / Melissa Valente-Paterno / Chelsea De Bellis / Phuong M Huynh / Zhe Fan / Edward M Marcotte / Dorota Wloga / Khanh Huy Bui /    Abstract: Cilia are ubiquitous eukaryotic organelles responsible for cellular motility and sensory functions. The ciliary axoneme is a microtubule-based cytoskeleton consisting of two central singlets and nine ...Cilia are ubiquitous eukaryotic organelles responsible for cellular motility and sensory functions. The ciliary axoneme is a microtubule-based cytoskeleton consisting of two central singlets and nine outer doublet microtubules. Cryo-electron microscopy-based studies have revealed a complex network inside the lumen of both tubules composed of microtubule-inner proteins (MIPs). However, the functions of most MIPs remain unknown. Here, we present single-particle cryo-EM-based analyses of the Tetrahymena thermophila native doublet microtubule and identify 42 MIPs. These data shed light on the evolutionarily conserved and diversified roles of MIPs. In addition, we identified MIPs potentially responsible for the assembly and stability of the doublet outer junction. Knockout of the evolutionarily conserved outer junction component CFAP77 moderately diminishes Tetrahymena swimming speed and beat frequency, indicating the important role of CFAP77 and outer junction stability in cilia beating generation and/or regulation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29685.map.gz emd_29685.map.gz | 456.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29685-v30.xml emd-29685-v30.xml emd-29685.xml emd-29685.xml | 75.7 KB 75.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29685_fsc.xml emd_29685_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_29685.png emd_29685.png | 182.7 KB | ||
| Masks |  emd_29685_msk_1.map emd_29685_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29685.cif.gz emd-29685.cif.gz | 16.9 KB | ||
| Others |  emd_29685_half_map_1.map.gz emd_29685_half_map_1.map.gz emd_29685_half_map_2.map.gz emd_29685_half_map_2.map.gz | 412.6 MB 412.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29685 http://ftp.pdbj.org/pub/emdb/structures/EMD-29685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29685 | HTTPS FTP |
-Related structure data
| Related structure data |  8g2zMC  8g3dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29685.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29685.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | composite map of 48-nm doublet from Tetrahymena strain CU428 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29685_msk_1.map emd_29685_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2 of 48-nm doublet from Tetrahymena strain CU428
| File | emd_29685_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 of 48-nm doublet from Tetrahymena strain CU428 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1 of 48-nm doublet from Tetrahymena strain CU428
| File | emd_29685_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 of 48-nm doublet from Tetrahymena strain CU428 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 48-nm repeat unit of microtubule doublet from Tetrahymena thermop...
| Entire | Name: 48-nm repeat unit of microtubule doublet from Tetrahymena thermophila strain CU428 |
|---|---|
| Components |
|
+Supramolecule #1: 48-nm repeat unit of microtubule doublet from Tetrahymena thermop...
| Supramolecule | Name: 48-nm repeat unit of microtubule doublet from Tetrahymena thermophila strain CU428 type: organelle_or_cellular_component / ID: 1 / Parent: 0 Macromolecule list: #46, #45, #38, #21, #16, #27, #15, #26, #24, #28, #17, #9, #18, #29, #33, #8, #14, #22, #20, #13, #23, #19, #10, #25, #34, #4, #7, #3, #32, #41, #39-#40, #2, #30, #6, #35, #11, ...Macromolecule list: #46, #45, #38, #21, #16, #27, #15, #26, #24, #28, #17, #9, #18, #29, #33, #8, #14, #22, #20, #13, #23, #19, #10, #25, #34, #4, #7, #3, #32, #41, #39-#40, #2, #30, #6, #35, #11, #36, #12, #31, #1, #5, #37, #42-#44 Details: Single Particle Analysis of the 48-nm repeating unit of the axoneme from intact cilia purified from Tetrahymena strain CU428 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.6 MDa |
+Macromolecule #1: RIB27A
| Macromolecule | Name: RIB27A / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.605641 KDa |
| Sequence | String: MSQYAYDFNP AKTQSLQSRP IEHNKFSAWT GDQLYRTSYG THWTSKPQEP KTHVPPGYAG YIPGLKPNNH YGASYGEIAK NCLSNPKVA QNPFKLASTG FNYQRHDFRD PSLTATTHKF GAQTLLKNHP SIDQKSNQWQ SQTHDSFRNP LHKPNPTYRE T DKDLQTQK ...String: MSQYAYDFNP AKTQSLQSRP IEHNKFSAWT GDQLYRTSYG THWTSKPQEP KTHVPPGYAG YIPGLKPNNH YGASYGEIAK NCLSNPKVA QNPFKLASTG FNYQRHDFRD PSLTATTHKF GAQTLLKNHP SIDQKSNQWQ SQTHDSFRNP LHKPNPTYRE T DKDLQTQK YFTKTSGFQQ NHTTFDRTGW VPEKVLHADR TTSEYRIHFN KQVPFHRDTV LFKERRLPPK EYNYKYMG UniProtKB: Uncharacterized protein |
+Macromolecule #2: RIB38
| Macromolecule | Name: RIB38 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.205637 KDa |
| Sequence | String: MAQACDKYTT ALMMGNWYEE RLAPNQPFRE NLQTKSVREE EAAISFASDT NKLIPLSRVN RNLPWDTKGV IADDGFKEFR SVNRTSYDP INLNNYYNLG DCRQLYKHYT AEKKYPENNP TQIQTLKANQ YSLLNQTNSQ HIHKERQVPN NVSDFGSTFK K FPETHEKF ...String: MAQACDKYTT ALMMGNWYEE RLAPNQPFRE NLQTKSVREE EAAISFASDT NKLIPLSRVN RNLPWDTKGV IADDGFKEFR SVNRTSYDP INLNNYYNLG DCRQLYKHYT AEKKYPENNP TQIQTLKANQ YSLLNQTNSQ HIHKERQVPN NVSDFGSTFK K FPETHEKF FGMTTYQQSF SRPLKETAHQ IIAQSEKKIN HFAGTNERPV HQQSTKMTSV LTAEKYKNEQ EPKDNTSIQR SW LPYKDKA LQVAENNLIK NQTLNQTKGF TPFNPLITYK NNMSQVQQHD IATSLPIGSG EHSDQKRTNQ PGEFRHVRTD VTL IRNKPV TKR UniProtKB: STOP protein |
+Macromolecule #3: CFAM166B
| Macromolecule | Name: CFAM166B / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.705826 KDa |
| Sequence | String: MYLQSKTTNN IPGYTGHIPQ EELHEDILQI NQAKSHIPGY AGYIPGIRSE NMYGKTYGKI TYMSVTNQHH KGPDLPPDLR YTSTVKGEF TDQKGVKKQE LLRQAVIEEA KITSSGYGSS PLRTKYIDPS EYKEKEIQPA PECNLTYQEA CKYAYSN UniProtKB: Ciliary microtubule inner protein 2A-C-like domain-containing protein |
+Macromolecule #4: CFAM166A
| Macromolecule | Name: CFAM166A / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.750811 KDa |
| Sequence | String: MFEGTRLSYV VPGYTGHIPK VIRDDPIPIQ KQEPKYQIPG YDGYIPSIKS ENIFGKTYGK ITYTISKGDQ HQGQDVNAQQ RYASVAEET YINQNQVLQR TAAEIVGVPP KQLQYTLTDK MKDSNFWQTQ LQHTKKLQQE ESQNVRPQQF EESLDKFYGD D KVDPIRIG ...String: MFEGTRLSYV VPGYTGHIPK VIRDDPIPIQ KQEPKYQIPG YDGYIPSIKS ENIFGKTYGK ITYTISKGDQ HQGQDVNAQQ RYASVAEET YINQNQVLQR TAAEIVGVPP KQLQYTLTDK MKDSNFWQTQ LQHTKKLQQE ESQNVRPQQF EESLDKFYGD D KVDPIRIG TPLPGYTGTN KRIVAANIFG QTFANARKTA AADQHNIDNE KMHNFKLQAQ NIPNIRR UniProtKB: Ciliary microtubule inner protein 2A-C-like domain-containing protein |
+Macromolecule #5: RIB22
| Macromolecule | Name: RIB22 / type: protein_or_peptide / ID: 5 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.401135 KDa |
| Sequence | String: MNSDLQNLIN RVRSALLSRN QNTIRGLARA FRQLDSYDRN NKVDRDEFFI VLKENGVSLT KQEQFVLHQC FDRNNEGTVD FDEFLCVIR GELNEQRKAI VLKAFQKFDP HNTGFVEIRD LRGVYNCDNH PKIKTGQMTE EQVFVEFLQN FCDNSKDGKI Q LKEWIDYY SAVSVSIQND DHFEQVVKIA WGV UniProtKB: EF hand protein |
+Macromolecule #6: CFAM166C
| Macromolecule | Name: CFAM166C / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.069318 KDa |
| Sequence | String: MNTTQMIPTY QDVKTGTTWL GGPTHVLQEQ HIPGYKGHVK GVQAESLHGK TFARITAECL NNRYKSGFIV DDEERFKTNY QLEYIRPNL RNNPTFTNNA SNVLNNRQRD DEVERLELIK AQLNSQPKTI EDVPPIDRVP VIGYQGFRPV YRHPLKQVKG P TEDEIDFQ ...String: MNTTQMIPTY QDVKTGTTWL GGPTHVLQEQ HIPGYKGHVK GVQAESLHGK TFARITAECL NNRYKSGFIV DDEERFKTNY QLEYIRPNL RNNPTFTNNA SNVLNNRQRD DEVERLELIK AQLNSQPKTI EDVPPIDRVP VIGYQGFRPV YRHPLKQVKG P TEDEIDFQ ARTILELTKG QKEAPKEIPI VGYTGFQKGI KAENMFGKTF KDLSNASNRI F UniProtKB: Uncharacterized protein |
+Macromolecule #7: CFAP107
| Macromolecule | Name: CFAP107 / type: protein_or_peptide / ID: 7 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.298023 KDa |
| Sequence | String: MNAQTIQDNV NKYRFGVLIG NFAEEKFGMD MAQRQIDERL PNSTMKDSYG LKNSALNCEP SKLTPIDKEF NQHVIFNTQG VQQHILFGH GVKQTDYNKR EYGTSYDLSF NQKIKPQTQI YSKYTPDALS ASRTFHKDTI FEKDYQKHIP ELGSKPTVPK D KARPYNEF TKTYDSTHMK IPLRK UniProtKB: Uncharacterized protein |
+Macromolecule #8: CFAP127
| Macromolecule | Name: CFAP127 / type: protein_or_peptide / ID: 8 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.558031 KDa |
| Sequence | String: MYPKQTQTSY KPQQHPAVTQ RQFIANDTYF NQQHYQRHEI EDALARELQR RKLDDEAKQR EIQKIYEESE ELKLLKEKIR MAQVNKERT RQIAENQLRK FQTKKVEAEL EENILRKCEE EKRYEAEKER ILNEQRMQGR FVLQQQMLEK RQAAEEAHQQ Y LLEKDQVN ...String: MYPKQTQTSY KPQQHPAVTQ RQFIANDTYF NQQHYQRHEI EDALARELQR RKLDDEAKQR EIQKIYEESE ELKLLKEKIR MAQVNKERT RQIAENQLRK FQTKKVEAEL EENILRKCEE EKRYEAEKER ILNEQRMQGR FVLQQQMLEK RQAAEEAHQQ Y LLEKDQVN AIVQNIINED KMAYESDKLK KKRNYQDMVE ALQEKAERKA QEKINEQLEN AKYKKFFEQK EQMQQEIKKK RE ELEAQKA IIFERLKAEE ERRRKEKELI EDLRMELYKE EFEAQERRKD REEREKRERT KQELQEAERQ HKYDKERQLA EEK RMEQQF REKMMEKYAQ DEKLEQLSAQ RRRMKEIDHK KEVERLWQEK LEMYREERER ELEEARKNEE LKSYKDELIR LEKE RLLRE YLDELAGFMP KGLLQTKEDT KFLKNQNPYA QRTGFASNFK IM UniProtKB: Meiosis-specific nuclear structural protein 1 |
+Macromolecule #9: CFAP161A
| Macromolecule | Name: CFAP161A / type: protein_or_peptide / ID: 9 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.489621 KDa |
| Sequence | String: MHNYNVDRKY QYNCRIGNWS EEWELDEYKM KEYLKNKEKQ KLVSTDKEDR LKRSLGGVNL SYANEGLVKF GYTVMIESDF MNAVVASNI WDKITGTEEA YGVTVTSQKE PIVRTAFQLL RYEQDQYQDD VIHYGQNFKI AVISRLSEKP LYLTSSHITP Q KCAKFSRK ...String: MHNYNVDRKY QYNCRIGNWS EEWELDEYKM KEYLKNKEKQ KLVSTDKEDR LKRSLGGVNL SYANEGLVKF GYTVMIESDF MNAVVASNI WDKITGTEEA YGVTVTSQKE PIVRTAFQLL RYEQDQYQDD VIHYGQNFKI AVISRLSEKP LYLTSSHITP Q KCAKFSRK QEVSISTNEG INSAWCFEYM DPKLRYEMKG QPVRSGEPVL IKHIPTCQWL ATDNVQYNNE FGQEFEVFCN SY QCHNKTQ NLIAEKVGRK TIETPMRNQT NQNTFRIVTA LYPSQETQQN FEASYSQTLP RRRVEINKAD LLNKLRRALI NKN GSNGLL FFKNELLKQD KDNSGYLPQQ QFKQTVQCHG INFSDLDIKE VIQSFQELDN KINYSELLIQ LKGTLSEKRR NLIN LAYDV LAEKLGQNVT LSGIFSAFNK IRHPDVVKKY GTDKEIFNNF VLSWGNLNPH IAISREQFES YYHDFSACVD KEDYF EVVL KNSWDIPN UniProtKB: EF hand protein, putative |
+Macromolecule #10: CFAP20
| Macromolecule | Name: CFAP20 / type: protein_or_peptide / ID: 10 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.976535 KDa |
| Sequence | String: MFKNTFQSGF LSILYSIGSK PLQIWDKSIR NGHIKRITDQ DILSSVLEIM GTNVSTNYIT APADPKETLG IKLPFLVMII KNLKKYYTF EVQVLDDKNV RRRFRASNYQ STTRVKPFIC TMPMRLDEGW NQIQFNLSDF TRRAYATNYI ETLRVQIHAN C RIRRIYFS ...String: MFKNTFQSGF LSILYSIGSK PLQIWDKSIR NGHIKRITDQ DILSSVLEIM GTNVSTNYIT APADPKETLG IKLPFLVMII KNLKKYYTF EVQVLDDKNV RRRFRASNYQ STTRVKPFIC TMPMRLDEGW NQIQFNLSDF TRRAYATNYI ETLRVQIHAN C RIRRIYFS DRLYSEEELP PEFKLFLPIQ GQNKTNV UniProtKB: Transcription factor iib protein, putative |
+Macromolecule #11: Parkin co-regulated protein PACRGA
| Macromolecule | Name: Parkin co-regulated protein PACRGA / type: protein_or_peptide / ID: 11 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.304816 KDa |
| Sequence | String: MLPKIPQQGN TVGSISKREN VASQLIVEDH LKNILAKSTN SNYKPWKIPQ APKNPHSPFG DFPKEYLPKS KLKSEQHAPV FEDSQAATV VNKFQGLRQG TGGKTSTQLP VKQPFQAPNP ICGAFKKRTI PVSEFRRYYD RGDLPIKVDH QGSVNKIIWV I QPDQLDYH ...String: MLPKIPQQGN TVGSISKREN VASQLIVEDH LKNILAKSTN SNYKPWKIPQ APKNPHSPFG DFPKEYLPKS KLKSEQHAPV FEDSQAATV VNKFQGLRQG TGGKTSTQLP VKQPFQAPNP ICGAFKKRTI PVSEFRRYYD RGDLPIKVDH QGSVNKIIWV I QPDQLDYH HYLPIFFDGL REKLDPYRFL AILGTYDLLE KGSNKILPVI PQLIIPVKTA LNTRDNEIIG IMLKILQRLV VS GDMIGEA LVPYYRQILP IFNLYKNCNK NVGDQIDYGQ RKKLTLGDLI QETLELFEQT GGEDAFINIK YMIPTYESCV HN UniProtKB: Parkin co-regulated protein |
+Macromolecule #12: IJ34
| Macromolecule | Name: IJ34 / type: protein_or_peptide / ID: 12 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.434062 KDa |
| Sequence | String: MNLDSIFEWI LKQPINYPGK HQDSQPRTNP AYKPLEAKAQ FHIIFKATGE YIREKMLDGK GVNMRDFGAF TFEIFSDTVK PAQHSNFDI AKDLDEQRAD RKHVHRIRPC FVPDKKFRYS LARYAGKEEI SAPKSQHSIY QKGFGMMFCN AGPIAASCYL G KDVVSSAH ...String: MNLDSIFEWI LKQPINYPGK HQDSQPRTNP AYKPLEAKAQ FHIIFKATGE YIREKMLDGK GVNMRDFGAF TFEIFSDTVK PAQHSNFDI AKDLDEQRAD RKHVHRIRPC FVPDKKFRYS LARYAGKEEI SAPKSQHSIY QKGFGMMFCN AGPIAASCYL G KDVVSSAH NAFIQAVSDL TTLGHGLNID FGFVKVYVQD RDLRYSYNQN FANTLNNKNF EYRLRKSDTP TSEHWNTTYE SK WAKSSLN SLLKRPNSNQ VRNYYEKTLA LKIMSLDLST AEQTNYSRIK PKPQQLPPLK K UniProtKB: Uncharacterized protein |
+Macromolecule #13: CFAP52A
| Macromolecule | Name: CFAP52A / type: protein_or_peptide / ID: 13 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73.429117 KDa |
| Sequence | String: MEKELDIQAV IGFTGKVNEG LILHPDNEHL IYPLGSTIVV RHIISRSQTF LRGHDNQISI ITVSKTGKYI ASGQKTYMGF QADIIIWDF ETRSLMHRLK LHKVLIQSLS FCCNEQYLAS LGGQDDKNMM IVWDVEQGKA LYGAPNRDVV NQIKFFNNSD D KIIAVLNN ...String: MEKELDIQAV IGFTGKVNEG LILHPDNEHL IYPLGSTIVV RHIISRSQTF LRGHDNQISI ITVSKTGKYI ASGQKTYMGF QADIIIWDF ETRSLMHRLK LHKVLIQSLS FCCNEQYLAS LGGQDDKNMM IVWDVEQGKA LYGAPNRDVV NQIKFFNNSD D KIIAVLNN GVQILTIDKA NKKVKSLDVN FGNIKRQYTC VAIDPSDTYC YCGTKTGDVF EINIERAIFK RLGPVKKLFS LG IGTLGLL PNGDIIVGAG DGTVAKLSIQ NMQVLSQSEV LGAVTSISFT GDYTHFFCGT SQSNIYWIDT EKLNPELRNT CHY ERINDI AFPYNYSDVF ATCSTTDIRV WNARNRQELL RIQVPNLECY CVTFMNDGKS IISGWNDGKI RAFLPQSGKL LYVI SDAHI HGVTALSTTS DCQRIVSGGS EGEVRVWVIN KQTQVMKASM KEHRGRVWSI QVKKNNDQAV SASADGSCII WDLKT FTRL MCLFESTLFK QVLYHPEESQ LLTTGSDRKI TYWEIYDGQA IRMLDGSEEG EVNALSITKE GEHFVSGGED KLVKLW GYD EGIKYYSGTG HSGAITRLQI SPDQRTVVSV GAEGAIFIWK MPEEVVHARA DNELPTISKE KHNNTQNQGD NKSSHSQ QH SQVSGASKGS QKRY UniProtKB: Cilia- and flagella-associated protein 52 |
+Macromolecule #14: DNA polymerase delta C4-type zinc-finger protein
| Macromolecule | Name: DNA polymerase delta C4-type zinc-finger protein / type: protein_or_peptide / ID: 14 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.493797 KDa |
| Sequence | String: MEEESIYNLI PKEYVPPPKE RRYRSQYPND LPPTGSTFIN HTTSRPGVAN LAGDFELSKG PHSHKDFCST FGRVKGSIKP STSDFRKKN TGTMGSNKLP IIKPFKYADH ESRRPNVPKI EEKPLMNQQS HKNYIVTNAV ENILSAPKAM KEPENYLTKR D YGKVPNYL ...String: MEEESIYNLI PKEYVPPPKE RRYRSQYPND LPPTGSTFIN HTTSRPGVAN LAGDFELSKG PHSHKDFCST FGRVKGSIKP STSDFRKKN TGTMGSNKLP IIKPFKYADH ESRRPNVPKI EEKPLMNQQS HKNYIVTNAV ENILSAPKAM KEPENYLTKR D YGKVPNYL VRNKEHLQTE YKMIQNLHLS EAEEQQRQRF LMSQEEVRQL RDGLKAKWEA VNKEYQTITH INKIDTVGLK RK KEECERE LTQLEKDIEK LNKQYIFVDT NM UniProtKB: DNA polymerase delta C4-type zinc-finger protein |
+Macromolecule #15: RIB43A protein
| Macromolecule | Name: RIB43A protein / type: protein_or_peptide / ID: 15 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.288318 KDa |
| Sequence | String: MKRVKESYFR EHSDWSDING CSGAKEFEGE DLAYDARIKY QKETQKQWIE QQIREKKMRE EAERNEERAY ATQTLELNRM RGMLEDDFN RKKASIRQAV KEENQQLDKQ KRDLEKQSNN EKLNYERTEI DMVKTRGQKR PFP UniProtKB: RIB43A protein |
+Macromolecule #16: RIB57
| Macromolecule | Name: RIB57 / type: protein_or_peptide / ID: 16 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.580914 KDa |
| Sequence | String: MDWINRLENI QNKIKFQLHA KKIDDLESLY RLMAEFDLDN SGRLEKDEFL KLLSKLGVFL TTQELRAVYD TYDTNRDEQI SYAEFVQMI RTTISEKRLA VVKHAFQFLD RRGNGKLAIQ DLVQQYRSEA HPRVRTRHKT AEQVKAEFVN AISKKSQDGQ T INENEFLE ...String: MDWINRLENI QNKIKFQLHA KKIDDLESLY RLMAEFDLDN SGRLEKDEFL KLLSKLGVFL TTQELRAVYD TYDTNRDEQI SYAEFVQMI RTTISEKRLA VVKHAFQFLD RRGNGKLAIQ DLVQQYRSEA HPRVRTRHKT AEQVKAEFVN AISKKSQDGQ T INENEFLE YYADVNATLP KEKEEYFIDI ILSTWGITSS VDYVSPERLN DLELILYEKI RQKTLPGEDE GRTVYRAFKY FD IAEKGVI DISQFKAALE KFGCVFDDKE IHALFSKFDV NRSGKICYDE FCGNFARLGS GNNPNVNPVF KLAREVPSPI INK LNADLK KQGPFAFRSL SKACQKADTF LNGSLSHDEF KWVIKEIGFS LTKTEFDNLF KFFDKNYEGK VIYSVFIDLV RGQQ SEIIT QSLSQGYDAL ASNNGGKVTV ESIVQRYNPL AIQIIYPTRK IAQDLQREFV AEWGGNLQQQ VNKNAFVEFF SDVRA FCDS ESKFHQYFSN AFGL UniProtKB: EF-hand pair protein |
+Macromolecule #17: CFAP182A
| Macromolecule | Name: CFAP182A / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.10984 KDa |
| Sequence | String: MDKLLASQRI NNSFGCKAMY QTTNNDYGQP FLQDNKTVPT YNELVEAQTE VTRAEKQLKN KTVATKFSDL KVEDEDEIMK EKRKFDRNV IVPKKFPLKE DSYKAPEPGL KIGNPLYMTA NREYGMLKPT AFEIPNRFYP KDNTYSKSFP GMHKFDGLNT N ITFSKVHR ALDEY UniProtKB: Uncharacterized protein |
+Macromolecule #18: CFAP143
| Macromolecule | Name: CFAP143 / type: protein_or_peptide / ID: 18 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.485744 KDa |
| Sequence | String: MELNQTTLES YNKQLQKGQG TLIGNWWEER ELRDVTGIGR SAHNYAKLKS TISQTGAQSL YKESPQTESV NDTNERTMGK KFNHIPNTT NSEYGKGFNK ADQLPRTGPV HLRTQQQMIN HIKQELNEIE NQKEIQRNIR YFQTTTQTEF GPKEHAMAGC T VGRRVMRT ...String: MELNQTTLES YNKQLQKGQG TLIGNWWEER ELRDVTGIGR SAHNYAKLKS TISQTGAQSL YKESPQTESV NDTNERTMGK KFNHIPNTT NSEYGKGFNK ADQLPRTGPV HLRTQQQMIN HIKQELNEIE NQKEIQRNIR YFQTTTQTEF GPKEHAMAGC T VGRRVMRT QNGQPITPDN RDEDILVDHG FLERQPFLTD EELKNQLPQG ESYLTQQPIT YWTEKTTDGF GCVYQSKSNP ND PKSTFKL NNQFLKTFHD YSHVRK UniProtKB: Uncharacterized protein |
+Macromolecule #19: CFAP21A
| Macromolecule | Name: CFAP21A / type: protein_or_peptide / ID: 19 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.502359 KDa |
| Sequence | String: MAANRTKDLF PGFKTAGAST KLPDESAMNC IKPKENLNPG TPEHIKKYRK SYKNQPGSTI LHYGIYDDQK PPETFVYGKK IEGSDHVQQ VMDSGKTDGI KQMINEIKEA KYHSRQVEPL GKRMERNYEF PQEVHQDNFK FGVATINSEN TAKEVMFPQK P AVNDQAAH ...String: MAANRTKDLF PGFKTAGAST KLPDESAMNC IKPKENLNPG TPEHIKKYRK SYKNQPGSTI LHYGIYDDQK PPETFVYGKK IEGSDHVQQ VMDSGKTDGI KQMINEIKEA KYHSRQVEPL GKRMERNYEF PQEVHQDNFK FGVATINSEN TAKEVMFPQK P AVNDQAAH DQYVRSHGNY EAGEQKNRNY NWKVDPQQHR FGKTDKIASN EVVYCLNQEA LQDQFPKTTI VQKKQEDFRN FK EDHLGLP KNLGQTNAKN NPDMVFGARL GGPDEWNAGK CISGEATLKE VQTDKDLGKT NRFGFRNITK DGDENRVFGV PTI RDDIQK PLMKSIADPN NYGDEKPAVN LLFPKKYDYM GVEQDDFKIK RHKYEIKDIF EKIGYKYKIG KFEGIFKRAQ EIEN STDNK VSCSSFLQAI QEMDHIE UniProtKB: EFHB C-terminal EF-hand domain-containing protein |
+Macromolecule #20: CFAP53
| Macromolecule | Name: CFAP53 / type: protein_or_peptide / ID: 20 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.717801 KDa |
| Sequence | String: MILSQNQQFN YRNPNERAIY QRRVQDEKLD RLKTKLNQEF YEKEFACWEN RGKAVNEGNY IKQRLAELRV RSQRALNERR QKLATLLGR EHEQYKQEIH DLQETPEDIR QKMIQRVNHL KEEKEKERQK IVEEKREQQF VRNADELRKV DADFHELKTY H EQNIQIME ...String: MILSQNQQFN YRNPNERAIY QRRVQDEKLD RLKTKLNQEF YEKEFACWEN RGKAVNEGNY IKQRLAELRV RSQRALNERR QKLATLLGR EHEQYKQEIH DLQETPEDIR QKMIQRVNHL KEEKEKERQK IVEEKREQQF VRNADELRKV DADFHELKTY H EQNIQIME KQKILQEKYE EEMIYAELYK YDIKKKEKEE LVKEYERKKK VDERNKVLHT QLAMNQTRRN QEKDLDSLEK MM LKEEWKK EEERHKKQQQ ELFQFNKDLN NEIKESNEEL QRRKAYEKDI EKQQDKEMIS RIITKEQMLD RLEAEQKRKF QEE TKAFLL NFKNRSNETH QYEAELERLI NEEAEKQWQR QNAIWQKERE AKIKLMHEVY DSRKTDIELK QTLVQVEKEQ KELE KDILT KAVAAYEQEV ERKKKELYLT NKQHQSSLIG QMHDKDRKRQ DELEKIRQEE EQERLRNIAY EQRLQEKKIQ GKQLL DELR KKRPF UniProtKB: Cilia- and flagella-associated protein 53 |
+Macromolecule #21: CFAP115
| Macromolecule | Name: CFAP115 / type: protein_or_peptide / ID: 21 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 110.670203 KDa |
| Sequence | String: MASVLAASGQ NILGAGRFNI SKNTITVNTD IEKLIKAEVF KKQIRIREFF YDFDRLRKGV VTEDKFRSAL SMLGMHLYEK DILSLINKY KSGPDQVKYT DFCDQIDAQF FDYNLAKENL ESTKSSSQIS QNESAIIQQI LLSIKQEIRT RRILLKQPFQ D FDRTRCSH ...String: MASVLAASGQ NILGAGRFNI SKNTITVNTD IEKLIKAEVF KKQIRIREFF YDFDRLRKGV VTEDKFRSAL SMLGMHLYEK DILSLINKY KSGPDQVKYT DFCDQIDAQF FDYNLAKENL ESTKSSSQIS QNESAIIQQI LLSIKQEIRT RRILLKQPFQ D FDRTRCSH ITVDQFSRVL TQLSILPNEQ LFNLLIRRYI DNGNPKEVNY VKFCDDVDNV SEMLETVIKG IKPNEKVFDP NE DIVEDTK DLKLMSTLFT SKKLNFGNKQ LNDVIVKIQA EVVMKRIRIR EFFKDYDNLR KGVVSEQQFR RVLDVSNIKL SEQ DINLLL SEFKVDNIPN GQVRYSDFCE VIDEIFTKKA IDKDPLATVT QFQPETTLPA RRYYLNLTEE EKGQLRHLLN LYHK EIQNK RVLIKPHFED FDKTKQGYIS KNQFLRILNQ FSLFPSEEAL NLILKRYTDK GTLDEVNYYE FCRDVDIYDE GVLVS KTYA DSFKNYQKPE KEFQPFIHND IPNDIEDLLA KLRAKVKEQR LRISEFLRDF DKLRSGNITK EQLRLGLSMA KLPLSD AEF TLVIENFAST TKENNVRWKD FCDQIDSVFT QKNLEKKSAT LPVFNPSTEY KYGRRGLTEQ ERRIAEHMKA RFRQFCQ AT RIDIKQFFQD WDRNGRNKVT PKQFRQVLTT VNFNISDPEF NAISRMYSSE DEHDVRYVDF IRDTKVYEQH LWSSNQTK D FGAHGLDTRK RPVDANVLLE EIKNIIKINR LRVGEYFADY DPLRKGVIPT NKFRGVISQM KIDLDEEQIL KLENMYILQ SDPTKLDYAS FLEDVNIVFT KTGLEKDPIA KPVPFEKKNA IDPRDVLTPQ EEEDLHVLLL RLEAVISKHR IMFKTHFQDK DIAKSGKVS FTRFRSILDL HKLPLTDQQF KLLCKRFAHQ GVEFNYIEFD EVVKKYENLY S UniProtKB: EF hand protein |
+Macromolecule #22: Nucleoside diphosphate kinase
| Macromolecule | Name: Nucleoside diphosphate kinase / type: protein_or_peptide / ID: 22 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.642727 KDa |
| Sequence | String: MADIRYIFIV EWFDTAASLI RTYYLTYFTQ DKTIEMYDLK NKKVFLKRCE YAIKDSDLYI GSILNVYSRQ LKIVDFADVF TRSKFQNIK EKTFAMIKPD AYIHIGKIIS IIERSGLQIS NLKMTKMSQE DAREFYGEHK GKPFYDGLVN FMSSDLIVGM E LVGDNAIK ...String: MADIRYIFIV EWFDTAASLI RTYYLTYFTQ DKTIEMYDLK NKKVFLKRCE YAIKDSDLYI GSILNVYSRQ LKIVDFADVF TRSKFQNIK EKTFAMIKPD AYIHIGKIIS IIERSGLQIS NLKMTKMSQE DAREFYGEHK GKPFYDGLVN FMSSDLIVGM E LVGDNAIK RWRELLGPTN TLVAREQAPN SIRGLFGTDG TRNACHGSDS PGSAFRELNF FFAKTSKLKT QAFFNQCTCC VI KPHIVKQ NQVGEVIDMI LSEGFEISAL QTFFLDRPTA EEFYEVYKGV LPEFNAIAEH LTSGMCYALE VRQENAVKSF RDI AGPHDP EIAKVIRPNT IRARFGIDRV KNGIHCTDLE DDGVLEVEYF FNALQN UniProtKB: Nucleoside diphosphate kinase |
+Macromolecule #23: Cilia- and flagella-associated protein 45
| Macromolecule | Name: Cilia- and flagella-associated protein 45 / type: protein_or_peptide / ID: 23 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.580438 KDa |
| Sequence | String: MPSSQASGGS KKSNADETLF GSRKTNKNNT KAQEEVVDKA RKGDFTHPEV VLIGESELQR MKNNAIITTK EETLHQKKIL EEQKEKQMA VSKAKKQKML QMEEEKKKQV PLSDFQLEEK VVKDSLLNRA QEIMNEQMDD VKEMNKMVMY AKCVTIRDKQ L TEKKDIRD ...String: MPSSQASGGS KKSNADETLF GSRKTNKNNT KAQEEVVDKA RKGDFTHPEV VLIGESELQR MKNNAIITTK EETLHQKKIL EEQKEKQMA VSKAKKQKML QMEEEKKKQV PLSDFQLEEK VVKDSLLNRA QEIMNEQMDD VKEMNKMVMY AKCVTIRDKQ L TEKKDIRD EKKLEEKRKD LMMEIERLKK IKYYEEQEKK KKEEQKEGHL KIIDQIKENE LVRLRAKEEQ EREGQVMIKA IK ELEVEEQ HKALKKKSDQ KKLLDEIFEA NQNAILVKQK RVQEEKVEEE KIVKYNLEKA QKEAEYIAEQ KRIKDEKEKE LQR LREMQE KANDRQAELD FLRAKRAMEQ NERNARDKER REAELRQKIN QELLEARRQQ QYEKEQRLQE QAKNDRDEFQ RIIQ AQKQE REIEIKLQIE REQQIKKHAD ELRKQIALNE EKIKQDQRNR LEEGKKIRDQ LQQQKKMLES IKSQKLEQLQ KMNIP QKYT SELERKKINI UniProtKB: Cilia- and flagella-associated protein 45 |
+Macromolecule #24: CFAP210
| Macromolecule | Name: CFAP210 / type: protein_or_peptide / ID: 24 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.707121 KDa |
| Sequence | String: MSVQSQSRQN KSSYGSQRQG SIQQQSINQQ SRTSNNFNQS NKTFTQKNTI QVPFSEIVRM KENCQINSDP EKERKLKEKT ELYELSKQR VKNWPNTIQA IRKKKDETRF EKFKKDEEER RRVDEEEARY QANVKKEIIE KANKQIYEGN DRVKAFQSKL L LVDTLQER ...String: MSVQSQSRQN KSSYGSQRQG SIQQQSINQQ SRTSNNFNQS NKTFTQKNTI QVPFSEIVRM KENCQINSDP EKERKLKEKT ELYELSKQR VKNWPNTIQA IRKKKDETRF EKFKKDEEER RRVDEEEARY QANVKKEIIE KANKQIYEGN DRVKAFQSKL L LVDTLQER EGQIELKKQV QEMEKMREEI HLENMKENIR KLEEEEQRKK EELEEKKRHA QKILREQHEE MKVKYIKRLQ DE KIEGEII KQRALEAIAK EKEEERARRE KAIQIQIDTK KGNEVLQTIR AELKEKEKLE DEKIKQYAEK KEKITEMRRL REE MRFKEK QEQRQKLIDE QIKRLAAIKN EEESRINKQI IEAQMKADEA EKKKREAIAA QKKRFDESIQ LTLKRKEQQV VEQK MEDQA YQQYWQKKNV QIQQREHQEK REAFNRAKDL QDFHKVQANE KQKLREEAFL NEMDEAEKIK QRLENDDKNF NSWAE RMIR EWNSDGKDIK PLLKELATYN KKL UniProtKB: Trichohyalin-plectin-homology domain-containing protein |
+Macromolecule #25: RIB72B
| Macromolecule | Name: RIB72B / type: protein_or_peptide / ID: 25 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.972898 KDa |
| Sequence | String: MASGFPKLPG FVPTQPIEQT SFKRVSAAKQ EYNRDYLHKN VPPVVYPLPR QVPKEPPSQK GMFNPNYMST THDTYRPVNC PNDVTELYQ PTWVKMDRQV LRFNAYFKES CVESSLEAYR VRKVVLFFYL EDSSIEISEP KQMNSGIPQG SFLKRQKVLK A DGSGVYLS ...String: MASGFPKLPG FVPTQPIEQT SFKRVSAAKQ EYNRDYLHKN VPPVVYPLPR QVPKEPPSQK GMFNPNYMST THDTYRPVNC PNDVTELYQ PTWVKMDRQV LRFNAYFKES CVESSLEAYR VRKVVLFFYL EDSSIEISEP KQMNSGIPQG SFLKRQKVLK A DGSGVYLS LYDFQIGQDL EFFGKIFHIY DCDIYTREFY ENLGIPQGAS ELPDSDNWEK KTLNKFVPVK DHMLKDFMEH KL GGGRVPS QKQFLENDRK VLKFFVFSEI PFFLHYYLAD DTLEIREIHR QNSGRDPFPL YLKRQKLPKQ FALNQPGQTY AEN FVKPTD IKFGEDLVAF GKKFRINGCD KYTQDYYREK YNIDFPLGED YEMQYRDIVK REIPPYNGFG DEEDSLGYVY RLIP KPPRK DFFKWVDNQV TLRFNAKFNT NKPEDIHRRF IITYYLNDDS LQVYEPAIRN SGIPDGKFLE KNRYKNVQTN NEYFQ PTDL VVGKDVIING YSFRILECDE YTLKWFANNI VQ UniProtKB: EF-hand protein |
+Macromolecule #26: Protofilament ribbon protein
| Macromolecule | Name: Protofilament ribbon protein / type: protein_or_peptide / ID: 26 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.224398 KDa |
| Sequence | String: MKELSQIIDK QISQLNLFGK IKKRKRQSNI YKMSGNTNSD FNRTNYQHKE QIIRCGISSL KCLDGEDLNQ GNRRRLQQLQ QRDWIEQQI REKEERKRQE DEEKKAFEQQ TLHINMMRGD LEDNLNQKRR NWEKNTKEFN IQQRNEKLDY ERSSHLDNQA Q NQYHITYC ...String: MKELSQIIDK QISQLNLFGK IKKRKRQSNI YKMSGNTNSD FNRTNYQHKE QIIRCGISSL KCLDGEDLNQ GNRRRLQQLQ QRDWIEQQI REKEERKRQE DEEKKAFEQQ TLHINMMRGD LEDNLNQKRR NWEKNTKEFN IQQRNEKLDY ERSSHLDNQA Q NQYHITYC NTNNFQTENT GTCTSAFGPH RVIPYHWKGM NPQQKKDIIL EQDQQRHERE ILKNLERDED KAFSNQTEHN RF MLINLER QKNRQHRQRM DEIKEFNLLA AKEQKIKLKH MYD UniProtKB: Protofilament ribbon protein |
+Macromolecule #27: RIB35
| Macromolecule | Name: RIB35 / type: protein_or_peptide / ID: 27 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.952652 KDa |
| Sequence | String: MSLYGVGLNV QVDRIKTEFR LYVSKQKGGL NVRNLRRIFE QLDYQQNGQL NLDDFKAGLN KFGFFLKNVD YQCLLKYFDV EQNGNVNYL AFLSCISQTL NQRRANFVLK VFDSIDSEQK GAVHPFSLAE RLEVVKNKDY QIGSKTREQL VGEYVAQFHK N QDGLITRD ...String: MSLYGVGLNV QVDRIKTEFR LYVSKQKGGL NVRNLRRIFE QLDYQQNGQL NLDDFKAGLN KFGFFLKNVD YQCLLKYFDV EQNGNVNYL AFLSCISQTL NQRRANFVLK VFDSIDSEQK GAVHPFSLAE RLEVVKNKDY QIGSKTREQL VGEYVAQFHK N QDGLITRD EFVSYYEDLG LGISSDQHFI EILQTQWGIK EDDDLAITKE EVKGIVKTIR QKLIQKTKGA NDEFLLRKLF KE FDISNSG FLTLDEMNAM LIKLEIPIQK RYLNAVFSQL DRNHSGVIEF EEFVYYIVND PYP UniProtKB: EF-hand pair protein |
+Macromolecule #28: CFAP182B
| Macromolecule | Name: CFAP182B / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.702059 KDa |
| Sequence | String: MNYNNLTQEQ VQEQIKKLEE EHQDFHKKHA NTFFKTTYKT YGGEKQVIDV PKHFGHDGRF TDHLLHATMY RNHSLNTTCD KERWMKNSK DWMEHLN UniProtKB: Uncharacterized protein |
+Macromolecule #29: Flagellar FliJ protein
| Macromolecule | Name: Flagellar FliJ protein / type: protein_or_peptide / ID: 29 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.098964 KDa |
| Sequence | String: MINIQRSIKL NEKYQQAEQY SKKLEGISRI TNWHETKVKY DNANQNKYAR KMIDQEMKYA LKEVKMVRNV KLKELYEQEA KQWEQELSQ KGLAIYKHKH UniProtKB: Flagellar FliJ protein |
+Macromolecule #30: RIB27B
| Macromolecule | Name: RIB27B / type: protein_or_peptide / ID: 30 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.270043 KDa |
| Sequence | String: MSQTENILDK KQNTTKQGID HSKFYEWSKQ FQYISSYQSN SSDKPNELRS DAIQGYKGFI PQIQSNNMFG QTFSRSARQV LASNVSNNP FKLSSTGHNS LRHNYADQSL NAFTHKFGAT TIQKNHPSLE SNLKQSLYKQ QFSKPVFSNE NNQDFNADDL N GNNNLSKK ...String: MSQTENILDK KQNTTKQGID HSKFYEWSKQ FQYISSYQSN SSDKPNELRS DAIQGYKGFI PQIQSNNMFG QTFSRSARQV LASNVSNNP FKLSSTGHNS LRHNYADQSL NAFTHKFGAT TIQKNHPSLE SNLKQSLYKQ QFSKPVFSNE NNQDFNADDL N GNNNLSKK TILSGFMKNN SIVGFKECLD SQLQLKDSKS EYNDKFQSKN RIHRDTSIFH IRNKSQIIIP A UniProtKB: Uncharacterized protein |
+Macromolecule #31: STPG2
| Macromolecule | Name: STPG2 / type: protein_or_peptide / ID: 31 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.647141 KDa |
| Sequence | String: MADKVEEEQA PPKIYTYIDL LKFENPEIYS SFGKMRLSHK TTAPKYGFGT ADRNKQAKVF QNKELAKTNF AGKSSPGPAY DVRDTDYFT YQKAPKWKIG SEVRNTLNTG SKHDFYLRKD VDFDPLEADI FRRPKAPTVR IGLELRFPND PKRHKGTPGP Q YNPTLRHE ...String: MADKVEEEQA PPKIYTYIDL LKFENPEIYS SFGKMRLSHK TTAPKYGFGT ADRNKQAKVF QNKELAKTNF AGKSSPGPAY DVRDTDYFT YQKAPKWKIG SEVRNTLNTG SKHDFYLRKD VDFDPLEADI FRRPKAPTVR IGLELRFPND PKRHKGTPGP Q YNPTLRHE IPNPPKFSFG FRREIQGFSP LVANSSTPQL VGPGSYIQKN VPNTSKIRNE PKWSFPNAER FSGFQADLSN AH YTKPRAI GTQYDSRKQT LPMFSFGKST RESKRGTFKD MMSTQEVRIR ISMPKF UniProtKB: Sperm-tail PG-rich repeat protein |
+Macromolecule #32: RIB26
| Macromolecule | Name: RIB26 / type: protein_or_peptide / ID: 32 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.114875 KDa |
| Sequence | String: MDDINKKVTT SKPSNVIFNT QTRGYCPGYL SPHNQISKKG LIVLQEWWGM NESICLLADE FAHQGFKVLV PDLYRGKVAK SHEEAGHLL TGLDWKGAIE DIAHALQYLI KLQGCTSVGI TGFCMGGALT LASLSAIEGF SCGSPFYGIC DQQTFPVTNI R VPVLAHFG ...String: MDDINKKVTT SKPSNVIFNT QTRGYCPGYL SPHNQISKKG LIVLQEWWGM NESICLLADE FAHQGFKVLV PDLYRGKVAK SHEEAGHLL TGLDWKGAIE DIAHALQYLI KLQGCTSVGI TGFCMGGALT LASLSAIEGF SCGSPFYGIC DQQTFPVTNI R VPVLAHFG EDDSMVGFSD VESAKALKLK AHEASVDFRL RTYPQAGHAF MRKESKTYQP QAAAAAFKET VEFFQQLLK UniProtKB: Dienelactone hydrolase family protein |
+Macromolecule #33: CFAP129
| Macromolecule | Name: CFAP129 / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.385977 KDa |
| Sequence | String: MISKVSGYQG FSSYGDKPYP SLKPGRQVLS PNDVWEQTQR TRDDASMYEN HQHYKTVYKV DVSNAVQPKV YQNHTHVKKG LQTQYQLQA TGKSVLSYGS DRKDQTFDPN NETSKLKTGV EHWKSNYNAN IKDPYSYSKA SRPEWSYHLK PHQVDSKIGP T EYKTTFGE ...String: MISKVSGYQG FSSYGDKPYP SLKPGRQVLS PNDVWEQTQR TRDDASMYEN HQHYKTVYKV DVSNAVQPKV YQNHTHVKKG LQTQYQLQA TGKSVLSYGS DRKDQTFDPN NETSKLKTGV EHWKSNYNAN IKDPYSYSKA SRPEWSYHLK PHQVDSKIGP T EYKTTFGE FGTKPTDKLN EYGNEIINKH EDPLKMGTTK STFHIPNYTG FIPAARTVGK SLEHAAALNS RIDKSRATIV DN YHTKIPG YAGHQPKAPV NQRGFLREHC FSTVSSLKL UniProtKB: Ptype ATPase, putative |
+Macromolecule #34: Flagellar microtubule protofilament ribbon protein
| Macromolecule | Name: Flagellar microtubule protofilament ribbon protein / type: protein_or_peptide / ID: 34 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.643695 KDa |
| Sequence | String: MNYNLPKTVP LLPGHCFPDH LKESHHKTQQ FTLVNNVHCE KTNYIEEKND PYLIDSLRMG TPPDLTYGKR KAPINDYIPR IQPPWLKYD RQVLRFYCYF QESVVENPYE NYRIRKCTLY YYLSDGTIHV NEPKIMNSGI EQGVFIKRQQ IPKQLNSTDY Y TWEDLNVG ...String: MNYNLPKTVP LLPGHCFPDH LKESHHKTQQ FTLVNNVHCE KTNYIEEKND PYLIDSLRMG TPPDLTYGKR KAPINDYIPR IQPPWLKYD RQVLRFYCYF QESVVENPYE NYRIRKCTLY YYLSDGTIHV NEPKIMNSGI EQGVFIKRQQ IPKQLNSTDY Y TWEDLNVG ININLFERVF RIVDCDSFTK EFFVYMGKKM NSPERVPNDA FEAQKTLKDI KIPPPNTKEY NEYNEVKLGG GH PNTGLQK YLENDRKVLS FNVLWNDTSL EGGLNYYILN YFLADDTTEI KEIKRHNSGK DAFPLFLCRK KLPKEPIMTH YPG MTLKKE EYYSPSDLVC GNMVRIYNKD CLIFNCDAYT KEWYLQNMNL QQIPITLKKE DIKRFYQPVP PYNGFGSEAD SLGS VYNLQ PKAPRKDINK MYTQDQYILR FEGKLISQNK EDNHRKFIIS FFCGDDTIMV YETADKNSGI WGGKFLERMN HNNPI NNKP YTELDFQIGE IIQLGVYRFQ LLRADEYTHK YMKSKPEVFK EADIEYTLNH LRKFATKYKS YDEFMVLLIK NIDPNR RGV IDFNDFVVGL RRLGYNLTYQ EIYTLMRYFD LDENWKLDVK SLFVALGGKQ UniProtKB: Flagellar microtugule protofilament ribbon protein |
+Macromolecule #35: Parkin co-regulated protein PACRGB
| Macromolecule | Name: Parkin co-regulated protein PACRGB / type: protein_or_peptide / ID: 35 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.41318 KDa |
| Sequence | String: MQSLSPSRKV KPNNTTHLLN ISSKFTGTGG NCSSLAPKKV QPIPKVKKTG IFEAKAIEAV EINLFKKYYD RGDLPIAVNF HGAHRKISW KLEPELLDYH LYLPIFFEGL RETEEPYKFL ADQGCEELLA RGGAKIFAVL PQLIIPIKKA LSTKSHPIMC T TLKKIQKL ...String: MQSLSPSRKV KPNNTTHLLN ISSKFTGTGG NCSSLAPKKV QPIPKVKKTG IFEAKAIEAV EINLFKKYYD RGDLPIAVNF HGAHRKISW KLEPELLDYH LYLPIFFEGL RETEEPYKFL ADQGCEELLA RGGAKIFAVL PQLIIPIKKA LSTKSHPIMC T TLKKIQKL VTSSDMIGEA LVPYYRQILP VMNMFKNKRL NIGDHIDYSQ RKNENLSDLI QETLEILEKY GGEDAYINIK YM IPTYESC MF UniProtKB: Parkin co-regulated protein |
+Macromolecule #36: OJ2
| Macromolecule | Name: OJ2 / type: protein_or_peptide / ID: 36 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.931387 KDa |
| Sequence | String: MPPIDTKKSQ ITDFSQSTRL QYLGDKKSQK SAAFFRDTRV SSCSYISLKG GVPRAIPYYG SPTKTYADTM SKSSYISSLN HDTYRVRPY QHVGMSQKLL EPYHPHSYRN RLPVPDAPPQ FSNASQIEVG DRSEVNHRRF VSQSKNVYGN FGKFDPVSNP G ILASKTKW HHHLQSK UniProtKB: Uncharacterized protein |
+Macromolecule #37: CFAP77A
| Macromolecule | Name: CFAP77A / type: protein_or_peptide / ID: 37 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.989633 KDa |
| Sequence | String: MSRTSTGFYP WNNRTQNTLL LKDDVGRSKP SAYSLPPVDY TYGRPLERDE EGAKEVSMTW KYHAETRPKE PDRDFAKLNK MSLTQGLHK SNQFSDYRKR VDARMKVVKG RNALPLYIPE EEFRYGLPNR PSTPIKLVLG NCYGLEFQQR IEQDYMYRSQ Q DSFKPRLL ...String: MSRTSTGFYP WNNRTQNTLL LKDDVGRSKP SAYSLPPVDY TYGRPLERDE EGAKEVSMTW KYHAETRPKE PDRDFAKLNK MSLTQGLHK SNQFSDYRKR VDARMKVVKG RNALPLYIPE EEFRYGLPNR PSTPIKLVLG NCYGLEFQQR IEQDYMYRSQ Q DSFKPRLL PKGNKASDLS YTYTKQKLSQ IQQNNNKELF KLTKFKKVQP RTNTYLHKNV TDVGKTSEIP NHEEHQHYDA NE VPNQAVE UniProtKB: Sperm-tail PG-rich repeat protein |
+Macromolecule #38: OJ3
| Macromolecule | Name: OJ3 / type: protein_or_peptide / ID: 38 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.31563 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #39: SB1
| Macromolecule | Name: SB1 / type: protein_or_peptide / ID: 39 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.793787 KDa |
| Sequence | String: MIRDFVLQQT TEPEKKFNST VLIGNWYEER CDPNREQSKF YNERKFADNN YQKYTLSENK FQDTSNSWLN FQENKPEVKN DQFITMNMQ EYKKPSEQKR NSELKPFIVK KSHFDKNPHE LEEYREKWTK SAHTFDRMYL GTQKPN UniProtKB: Uncharacterized protein |
+Macromolecule #40: STPG1A
| Macromolecule | Name: STPG1A / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.132254 KDa |
| Sequence | String: MQCLLLVFQF NQNYTVKKIA EKEILSNFYL QSRLIDQQNS SYLNMSQSLP SVQKSASMTL MPIMEMYNIS TRQHAAWGLD GYEVPKKYF DHLKVVQDRH FEEISKSGKA TKNNKIITKR GSYLEDEIKF RGQNPGPQKY DVTYKWVSDA EIEKGKKLPK N TKKNTFIE ...String: MQCLLLVFQF NQNYTVKKIA EKEILSNFYL QSRLIDQQNS SYLNMSQSLP SVQKSASMTL MPIMEMYNIS TRQHAAWGLD GYEVPKKYF DHLKVVQDRH FEEISKSGKA TKNNKIITKR GSYLEDEIKF RGQNPGPQKY DVTYKWVSDA EIEKGKKLPK N TKKNTFIE QIFLEQQRRG IPGPGKYNIL KTDEQVKAEA EKMNKKLKYG ERSNYLQEYE YLSSTLPGPG NYNPRPILPK IH KDNMSPD KWIAFHKAKL SKTAKSSLPD VGTYKMNYPL DYATFGKMLV KTQEEGGNKK SSVRYMGTEE RFKDPKKTKS KTS QIVPGP GQYPLVAKWQ GKEQKKDSKD KNWMDSITTG ISKSIYYS UniProtKB: Sperm-tail PG-rich repeat protein |
+Macromolecule #41: Nebulin
| Macromolecule | Name: Nebulin / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.047602 KDa |
| Sequence | String: MTDNPQQPHK SKQEIQREQR KELARELRKA HFDLGFKEGF DDETRYREFY KWYDLEQSEK TKQEMLKLRN DLRSTHYILG TDDPNKLFV STATQSFVKP VNPQVSQLSV ETKNDLRSHH FNLGHYNDKV LSDYKLNYDQ KQIDPETLKD RKEQINFLRK H NHDFGDKN ...String: MTDNPQQPHK SKQEIQREQR KELARELRKA HFDLGFKEGF DDETRYREFY KWYDLEQSEK TKQEMLKLRN DLRSTHYILG TDDPNKLFV STATQSFVKP VNPQVSQLSV ETKNDLRSHH FNLGHYNDKV LSDYKLNYDQ KQIDPETLKD RKEQINFLRK H NHDFGDKN NYHSSMYNEN FNKSYDPHFL KQGKSKEEIH QQIVDLRKTN LVMGNNNPQF TSEAMSEFNN KPQAFRTQVD LG LKKSHFK LGEDPSLYET TTAKTYQGKQ MFQHDPEKIK ALSKDLRAEH FKLGNDPQSY TSEAAAKFKE FDKNSLTQQP DMS YLYRSH FNLEGFGGSN PVQHYVSNYK QNYEPKAAQK SEATRNDRAD RGSHIVFGSD KIDDQFKSEA QKNFVNFGRQ APSA LEKEV QADLRRHHYQ FGTDQPEMIS EMKKTFNDKT KESSQSKLDP NLIKDLRSNH FEYGTMGNEY TTTMQDIGRY QCQPS RLNP ELAKDLRSHH FRPGDLEKYY DTTYRLAFID FKAV UniProtKB: Uncharacterized protein |
+Macromolecule #42: B5B6_fMIP
| Macromolecule | Name: B5B6_fMIP / type: protein_or_peptide / ID: 42 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.166234 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #43: CFAP112A
| Macromolecule | Name: CFAP112A / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.038562 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
+Macromolecule #44: B2B3_fMIP
| Macromolecule | Name: B2B3_fMIP / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.099145 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)AYA AA AYAYAAA AYAAAAYAAA AAAAAAAAAY AYAYAAAYAA AAAYAAAAAA YAAAAAAAYY AYAAAAYAAA AAAAAAAAAA AAA AAAAAY AAAYYYYAAA AAYAAYAAAY AAAAAAAYAA AYAAAAAAAA AAAAAAAAAA AAAAAYYAAA AAYAAAYAAA AAAA AAAAY AAAAAAYA(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
+Macromolecule #45: Tubulin alpha chain
| Macromolecule | Name: Tubulin alpha chain / type: protein_or_peptide / ID: 45 / Number of copies: 161 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.639035 KDa |
| Sequence | String: MREVISIHVG QGGIQVGNAC WELFCLEHGI QPDGQMPSDK TIGGGDDAFN TFFSETGAGK HVPRAVFLDL EPTVIDEVRT GTYRQLFHP EQLISGKEDA ANNFARGHYT IGKEIVDLCL DRIRKLADNC TGLQGFLVFN SVGGGTGSGL GSLLLERLSV D YGKKSKLG ...String: MREVISIHVG QGGIQVGNAC WELFCLEHGI QPDGQMPSDK TIGGGDDAFN TFFSETGAGK HVPRAVFLDL EPTVIDEVRT GTYRQLFHP EQLISGKEDA ANNFARGHYT IGKEIVDLCL DRIRKLADNC TGLQGFLVFN SVGGGTGSGL GSLLLERLSV D YGKKSKLG FTIYPSPQVS TAVVEPYNSI LSTHSLLEHT DVAVMLDNEA IYDICRRNLD IERPTYTNLN RLIAQVISSL TA SLRFDGA LNVDITEFQT NLVPYPRIHF MLSSYAPIIS AEKAYHEQLS VAEITNSAFE PANMMAKCDP RHGKYMACSM MYR GDVVPK DVNASIATIK TKRTIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVMRAVC MISNSTAIAE VFSRLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDL AALEKDYEEV GIETAEGEGE EEGY UniProtKB: Tubulin alpha chain |
+Macromolecule #46: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 46 / Number of copies: 161 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.617676 KDa |
| Sequence | String: MREIVHIQGG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLERINVY YNEATGGRYV PRAILMDLEP GTMDSVRAGP FGQLFRPDN FVFGQTGAGN NWAKGHYTEG AELIDSVLDV VRKEAEGCDC LQGFQITHSL GGGTGSGMGT LLISKVREEY P DRIMETFS ...String: MREIVHIQGG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLERINVY YNEATGGRYV PRAILMDLEP GTMDSVRAGP FGQLFRPDN FVFGQTGAGN NWAKGHYTEG AELIDSVLDV VRKEAEGCDC LQGFQITHSL GGGTGSGMGT LLISKVREEY P DRIMETFS VVPSPKVSDT VVEPYNATLS VHQLVENADE CMVIDNEALY DICFRTLKLT TPTYGDLNHL VSAAMSGVTC CL RFPGQLN SDLRKLAVNL IPFPRLHFFM IGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMCAADPRH GRYLTASALF RGR MSTKEV DEQMLNVQNK NSSYFVEWIP NNIKSSICDI PPKGLKMAVT FVGNSTAIQE MFKRVAEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATAEEEGE FEEEEGEN UniProtKB: Tubulin beta chain |
+Macromolecule #47: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 47 / Number of copies: 161 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
+Macromolecule #48: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 48 / Number of copies: 161 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
+Macromolecule #49: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 49 / Number of copies: 161 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Concentration | 2.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)