+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of ACLY-D1026A-products-asym | |||||||||
 Map data Map data | Structure of ACLY-D1026A-products-asym | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mutant / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP citrate synthase / ATP citrate synthase activity / citrate metabolic process / Fatty acyl-CoA biosynthesis / acetyl-CoA biosynthetic process / ChREBP activates metabolic gene expression / coenzyme A metabolic process / oxaloacetate metabolic process / negative regulation of ferroptosis / cholesterol biosynthetic process ...ATP citrate synthase / ATP citrate synthase activity / citrate metabolic process / Fatty acyl-CoA biosynthesis / acetyl-CoA biosynthetic process / ChREBP activates metabolic gene expression / coenzyme A metabolic process / oxaloacetate metabolic process / negative regulation of ferroptosis / cholesterol biosynthetic process / lipid biosynthetic process / azurophil granule lumen / fatty acid biosynthetic process / ficolin-1-rich granule lumen / ciliary basal body / Neutrophil degranulation / extracellular exosome / extracellular region / nucleoplasm / ATP binding / metal ion binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Wei X / Marmorstein R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Allosteric role of the citrate synthase homology domain of ATP citrate lyase. Authors: Xuepeng Wei / Kollin Schultz / Hannah L Pepper / Emily Megill / Austin Vogt / Nathaniel W Snyder / Ronen Marmorstein /   Abstract: ATP citrate lyase (ACLY) is the predominant nucleocytosolic source of acetyl-CoA and is aberrantly regulated in many diseases making it an attractive therapeutic target. Structural studies of ACLY ...ATP citrate lyase (ACLY) is the predominant nucleocytosolic source of acetyl-CoA and is aberrantly regulated in many diseases making it an attractive therapeutic target. Structural studies of ACLY reveal a central homotetrameric core citrate synthase homology (CSH) module flanked by acyl-CoA synthetase homology (ASH) domains, with ATP and citrate binding the ASH domain and CoA binding the ASH-CSH interface to produce acetyl-CoA and oxaloacetate products. The specific catalytic role of the CSH module and an essential D1026A residue contained within it has been a matter of debate. Here, we report biochemical and structural analysis of an ACLY-D1026A mutant demonstrating that this mutant traps a (3S)-citryl-CoA intermediate in the ASH domain in a configuration that is incompatible with the formation of acetyl-CoA, is able to convert acetyl-CoA and OAA to (3S)-citryl-CoA in the ASH domain, and can load CoA and unload acetyl-CoA in the CSH module. Together, this data support an allosteric role for the CSH module in ACLY catalysis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29668.map.gz emd_29668.map.gz | 37.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29668-v30.xml emd-29668-v30.xml emd-29668.xml emd-29668.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29668.png emd_29668.png | 177.5 KB | ||
| Filedesc metadata |  emd-29668.cif.gz emd-29668.cif.gz | 6.4 KB | ||
| Others |  emd_29668_half_map_1.map.gz emd_29668_half_map_1.map.gz emd_29668_half_map_2.map.gz emd_29668_half_map_2.map.gz | 37.7 MB 37.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29668 http://ftp.pdbj.org/pub/emdb/structures/EMD-29668 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29668 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29668 | HTTPS FTP |
-Validation report
| Summary document |  emd_29668_validation.pdf.gz emd_29668_validation.pdf.gz | 994.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29668_full_validation.pdf.gz emd_29668_full_validation.pdf.gz | 994.3 KB | Display | |
| Data in XML |  emd_29668_validation.xml.gz emd_29668_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_29668_validation.cif.gz emd_29668_validation.cif.gz | 13.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29668 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29668 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29668 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29668 | HTTPS FTP |
-Related structure data
| Related structure data |  8g1eMC  7rigC  7rkzC  7rmpC  8g1fC  8g5cC  8g5dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29668.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29668.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of ACLY-D1026A-products-asym | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_29668_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_29668_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ACLY D1026A mutant in complex with CoA and citrate
| Entire | Name: ACLY D1026A mutant in complex with CoA and citrate |
|---|---|
| Components |
|
-Supramolecule #1: ACLY D1026A mutant in complex with CoA and citrate
| Supramolecule | Name: ACLY D1026A mutant in complex with CoA and citrate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 480 KDa |
-Macromolecule #1: ATP-citrate synthase
| Macromolecule | Name: ATP-citrate synthase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: ATP citrate synthase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120.940125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAKAISEQT GKELLYKFIC TTSAIQNRFK YARVTPDTDW ARLLQDHPWL LSQNLVVKPD QLIKRRGKLG LVGVNLTLDG VKSWLKPRL GQEATVGKAT GFLKNFLIEP FVPHSQAEEF YVCIYATREG DYVLFHHEGG VDVGDVDAKA QKLLVGVDEK L NPEDIKKH ...String: MSAKAISEQT GKELLYKFIC TTSAIQNRFK YARVTPDTDW ARLLQDHPWL LSQNLVVKPD QLIKRRGKLG LVGVNLTLDG VKSWLKPRL GQEATVGKAT GFLKNFLIEP FVPHSQAEEF YVCIYATREG DYVLFHHEGG VDVGDVDAKA QKLLVGVDEK L NPEDIKKH LLVHAPEDKK EILASFISGL FNFYEDLYFT YLEINPLVVT KDGVYVLDLA AKVDATADYI CKVKWGDIEF PP PFGREAY PEEAYIADLD AKSGASLKLT LLNPKGRIWT MVAGGGASVV YSDTICDLGG VNELANYGEY SGAPSEQQTY DYA KTILSL MTREKHPDGK ILIIGGSIAN FTNVAATFKG IVRAIRDYQG PLKEHEVTIF VRRGGPNYQE GLRVMGEVGK TTGI PIHVF GTETHMTAIV GMALGHRPIP NQPPTAAHTA NFLLNASGST STPAPSRTAS FSESRADEVA PAKKAKPAMP QDSVP SPRS LQGKSTTLFS RHTKAIVWGM QTRAVQGMLD FDYVCSRDEP SVAAMVYPFT GDHKQKFYWG HKEILIPVFK NMADAM RKH PEVDVLINFA SLRSAYDSTM ETMNYAQIRT IAIIAEGIPE ALTRKLIKKA DQKGVTIIGP ATVGGIKPGC FKIGNTG GM LDNILASKLY RPGSVAYVSR SGGMSNELNN IISRTTDGVY EGVAIGGDRY PGSTFMDHVL RYQDTPGVKM IVVLGEIG G TEEYKICRGI KEGRLTKPIV CWCIGTCATM FSSEVQFGHA GACANQASET AVAKNQALKE AGVFVPRSFD ELGEIIQSV YEDLVANGVI VPAQEVPPPT VPMDYSWARE LGLIRKPASF MTSICDERGQ ELIYAGMPIT EVFKEEMGIG GVLGLLWFQK RLPKYSCQF IEMCLMVTAD HGPAVSGAHN TIICARAGKD LVSSLTSGLL TIGDRFGGAL DAAAKMFSKA FDSGIIPMEF V NKMKKEGK LIMGIGHRVK SINNPDMRVQ ILKDYVRQHF PATPLLDYAL EVEKITTSKK PNLILNVAGL IGVAFVDMLR NC GSFTREE ADEYIDIGAL NGIFVLGRSM GFIGHYLDQK RLKQGLYRHP WDDISYVLPE HMSM UniProtKB: ATP-citrate synthase |
-Macromolecule #2: ACETYL COENZYME *A
| Macromolecule | Name: ACETYL COENZYME *A / type: ligand / ID: 2 / Number of copies: 4 / Formula: ACO |
|---|---|
| Molecular weight | Theoretical: 809.571 Da |
| Chemical component information |  ChemComp-ACO: |
-Macromolecule #3: OXALOACETATE ION
| Macromolecule | Name: OXALOACETATE ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: OAA |
|---|---|
| Molecular weight | Theoretical: 131.064 Da |
| Chemical component information |  ChemComp-OAA: |
-Macromolecule #4: UNKNOWN LIGAND
| Macromolecule | Name: UNKNOWN LIGAND / type: ligand / ID: 4 / Number of copies: 2 / Formula: UNL |
|---|---|
| Chemical component information | 
ChemComp-UNL: |
-Macromolecule #5: (3S)-citryl-Coenzyme A
| Macromolecule | Name: (3S)-citryl-Coenzyme A / type: ligand / ID: 5 / Number of copies: 3 / Formula: Q5B |
|---|---|
| Molecular weight | Theoretical: 941.642 Da |
| Chemical component information | 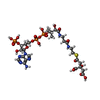 ChemComp-Q5B: |
-Macromolecule #6: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 6 / Number of copies: 3 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #7: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 90 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.50 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 183036 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|---|
| Output model |  PDB-8g1e: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)