+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of nuclear GAPDH under 8h Oxidative Stress | |||||||||
 Map data Map data | GAPDH,Nuclear Oxidation 8h,D2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GAPDH / energy / cytosolic protein / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidyl-cysteine S-trans-nitrosylation / Transferases; Transferring nitrogenous groups; Transferring other nitrogenous groups / negative regulation of endopeptidase activity / glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) / killing by host of symbiont cells / aspartic-type endopeptidase inhibitor activity / glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity / Gluconeogenesis / Glycolysis / GAIT complex ...peptidyl-cysteine S-trans-nitrosylation / Transferases; Transferring nitrogenous groups; Transferring other nitrogenous groups / negative regulation of endopeptidase activity / glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) / killing by host of symbiont cells / aspartic-type endopeptidase inhibitor activity / glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity / Gluconeogenesis / Glycolysis / GAIT complex / peptidyl-cysteine S-nitrosylase activity / positive regulation of type I interferon production / regulation of macroautophagy / defense response to fungus / lipid droplet / positive regulation of cytokine production / glycolytic process / cellular response to type II interferon / microtubule cytoskeleton organization / glucose metabolic process / NAD binding / antimicrobial humoral immune response mediated by antimicrobial peptide / disordered domain specific binding / NADP binding / microtubule cytoskeleton / neuron apoptotic process / nuclear membrane / microtubule binding / vesicle / killing of cells of another organism / positive regulation of canonical NF-kappaB signal transduction / negative regulation of translation / protein stabilization / ribonucleoprotein complex / intracellular membrane-bounded organelle / perinuclear region of cytoplasm / extracellular exosome / identical protein binding / nucleus / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo (humans) / Homo (humans) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.17 Å | |||||||||
 Authors Authors | Choi WY / Wu H / Cheng YF / Manglik A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Efficient tagging of endogenous proteins in human cell lines for structural studies by single-particle cryo-EM. Authors: Wooyoung Choi / Hao Wu / Klaus Yserentant / Bo Huang / Yifan Cheng /  Abstract: CRISPR/Cas9-based genome engineering has revolutionized our ability to manipulate biological systems, particularly in higher organisms. Here, we designed a set of homology-directed repair donor ...CRISPR/Cas9-based genome engineering has revolutionized our ability to manipulate biological systems, particularly in higher organisms. Here, we designed a set of homology-directed repair donor templates that enable efficient tagging of endogenous proteins with affinity tags by transient transfection and selection of genome-edited cells in various human cell lines. Combined with technological advancements in single-particle cryogenic electron microscopy, this strategy allows efficient structural studies of endogenous proteins captured in their native cellular environment and during different cellular processes. We demonstrated this strategy by tagging six different human proteins in both HEK293T and Jurkat cells. Moreover, analysis of endogenous glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in HEK293T cells allowed us to follow its behavior spatially and temporally in response to prolonged oxidative stress, correlating the increased number of oxidation-induced inactive catalytic sites in GAPDH with its translocation from cytosol to nucleus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29659.map.gz emd_29659.map.gz | 39.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29659-v30.xml emd-29659-v30.xml emd-29659.xml emd-29659.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29659.png emd_29659.png | 61.4 KB | ||
| Filedesc metadata |  emd-29659.cif.gz emd-29659.cif.gz | 5.6 KB | ||
| Others |  emd_29659_additional_1.map.gz emd_29659_additional_1.map.gz emd_29659_additional_2.map.gz emd_29659_additional_2.map.gz emd_29659_additional_3.map.gz emd_29659_additional_3.map.gz emd_29659_additional_4.map.gz emd_29659_additional_4.map.gz emd_29659_additional_5.map.gz emd_29659_additional_5.map.gz emd_29659_half_map_1.map.gz emd_29659_half_map_1.map.gz emd_29659_half_map_2.map.gz emd_29659_half_map_2.map.gz | 1.3 MB 1.3 MB 1.3 MB 1.3 MB 1.3 MB 39.7 MB 39.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29659 http://ftp.pdbj.org/pub/emdb/structures/EMD-29659 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29659 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29659 | HTTPS FTP |
-Validation report
| Summary document |  emd_29659_validation.pdf.gz emd_29659_validation.pdf.gz | 749.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29659_full_validation.pdf.gz emd_29659_full_validation.pdf.gz | 748.7 KB | Display | |
| Data in XML |  emd_29659_validation.xml.gz emd_29659_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_29659_validation.cif.gz emd_29659_validation.cif.gz | 13.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29659 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29659 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29659 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29659 | HTTPS FTP |
-Related structure data
| Related structure data |  8g12MC  8g13C  8g14C  8g15C  8g16C  8g17C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29659.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29659.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAPDH,Nuclear Oxidation 8h,D2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
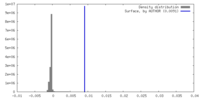
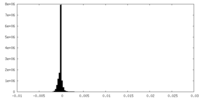
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: GAPDH,Nuclear Oxidation 8h,single subunit analysis,inactive
| File | emd_29659_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAPDH,Nuclear Oxidation 8h,single subunit analysis,inactive | ||||||||||||
| Projections & Slices |
| ||||||||||||
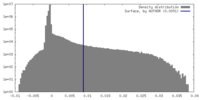
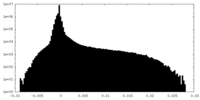
| Density Histograms |
-Additional map: GAPDH,Nuclear Oxidation 8h,single subunit analysis,inactive
| File | emd_29659_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAPDH,Nuclear Oxidation 8h,single subunit analysis,inactive | ||||||||||||
| Projections & Slices |
| ||||||||||||
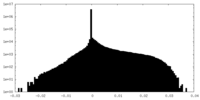
| Density Histograms |
-Additional map: GAPDH,Nuclear Oxidation 8h,single subunit analysis,non defined
| File | emd_29659_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAPDH,Nuclear Oxidation 8h,single subunit analysis,non defined | ||||||||||||
| Projections & Slices |
| ||||||||||||
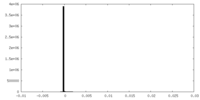
| Density Histograms |
-Additional map: GAPDH,Nuclear Oxidation 8h,single subunit analysis,non defined
| File | emd_29659_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAPDH,Nuclear Oxidation 8h,single subunit analysis,non defined | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: GAPDH,Nuclear Oxidation 8h,single subunit analysis,non defined
| File | emd_29659_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAPDH,Nuclear Oxidation 8h,single subunit analysis,non defined | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: GAPDH,Nuclear Oxidation 8h,D2,halfmap1
| File | emd_29659_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAPDH,Nuclear Oxidation 8h,D2,halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: GAPDH,Nuclear Oxidation 8h,D2,halfmap2
| File | emd_29659_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAPDH,Nuclear Oxidation 8h,D2,halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GAPDH
| Entire | Name: GAPDH |
|---|---|
| Components |
|
-Supramolecule #1: GAPDH
| Supramolecule | Name: GAPDH / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo (humans) Homo (humans) |
-Macromolecule #1: Glyceraldehyde-3-phosphate dehydrogenase
| Macromolecule | Name: Glyceraldehyde-3-phosphate dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO EC number: glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.983969 KDa |
| Recombinant expression | Organism:  Homo (humans) Homo (humans) |
| Sequence | String: GKVKVGVNGF GRIGRLVTRA AFNSGKVDIV AINDPFIDLN YMVYMFQYDS THGKFHGTVK AENGKLVING NPITIFQERD PSKIKWGDA GAEYVVESTG VFTTMEKAGA HLQGGAKRVI ISAPSADAPM FVMGVNHEKY DNSLKIISNA S(CSO)TTNCL AP LAKVIHDNFG ...String: GKVKVGVNGF GRIGRLVTRA AFNSGKVDIV AINDPFIDLN YMVYMFQYDS THGKFHGTVK AENGKLVING NPITIFQERD PSKIKWGDA GAEYVVESTG VFTTMEKAGA HLQGGAKRVI ISAPSADAPM FVMGVNHEKY DNSLKIISNA S(CSO)TTNCL AP LAKVIHDNFG IVEGLMTTVH AITATQKTVD GPSGKLWRDG RGALQNIIPA STGAAKAVGK VIPELNGKLT GMAFRVPT A NVSVVDLTCR LEKPAKYDDI KKVVKQASEG PLKGILGYTE HQVVSSDFNS DTHSSTFDAG AGIALNDHFV KLISWYDNE FGYSNRVVDL MAHMASKE UniProtKB: Glyceraldehyde-3-phosphate dehydrogenase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 45.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.17 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 764164 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)