+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Body1 of G4 RNA-mediated PRC2 dimer from multibody refinement | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  PRC2 / G-quadruplex RNA / RNP complex / chromatin modifier / PRC2 / G-quadruplex RNA / RNP complex / chromatin modifier /  GENE REGULATION GENE REGULATION | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
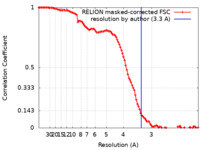
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.3 Å cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Jiarui JS / Vignesh VK | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structural basis for inactivation of PRC2 by G-quadruplex RNA. Authors: Jiarui Song / Anne R Gooding / Wayne O Hemphill / Brittney D Love / Anne Robertson / Liqi Yao / Leonard I Zon / Trista E North / Vignesh Kasinath / Thomas R Cech /  Abstract: Polycomb repressive complex 2 (PRC2) silences genes through trimethylation of histone H3K27. PRC2 associates with numerous precursor messenger RNAs (pre-mRNAs) and long noncoding RNAs (lncRNAs) with ...Polycomb repressive complex 2 (PRC2) silences genes through trimethylation of histone H3K27. PRC2 associates with numerous precursor messenger RNAs (pre-mRNAs) and long noncoding RNAs (lncRNAs) with a binding preference for G-quadruplex RNA. In this work, we present a 3.3-Å-resolution cryo-electron microscopy structure of PRC2 bound to a G-quadruplex RNA. Notably, RNA mediates the dimerization of PRC2 by binding both protomers and inducing a protein interface composed of two copies of the catalytic subunit EZH2, thereby blocking nucleosome DNA interaction and histone H3 tail accessibility. Furthermore, an RNA-binding loop of EZH2 facilitates the handoff between RNA and DNA, another activity implicated in PRC2 regulation by RNA. We identified a gain-of-function mutation in this loop that activates PRC2 in zebrafish. Our results reveal mechanisms for RNA-mediated regulation of a chromatin-modifying enzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29647.map.gz emd_29647.map.gz | 57.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29647-v30.xml emd-29647-v30.xml emd-29647.xml emd-29647.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29647_fsc.xml emd_29647_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_29647.png emd_29647.png | 79.6 KB | ||
| Masks |  emd_29647_msk_1.map emd_29647_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29647.cif.gz emd-29647.cif.gz | 8.4 KB | ||
| Others |  emd_29647_additional_1.map.gz emd_29647_additional_1.map.gz emd_29647_half_map_1.map.gz emd_29647_half_map_1.map.gz emd_29647_half_map_2.map.gz emd_29647_half_map_2.map.gz | 9.9 MB 58.3 MB 58.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29647 http://ftp.pdbj.org/pub/emdb/structures/EMD-29647 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29647 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29647 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29647.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29647.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.06344 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29647_msk_1.map emd_29647_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_29647_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29647_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29647_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : G4 RNA-mediated dimer of polycomb repressive complex 2
| Entire | Name: G4 RNA-mediated dimer of polycomb repressive complex 2 |
|---|---|
| Components |
|
-Supramolecule #1: G4 RNA-mediated dimer of polycomb repressive complex 2
| Supramolecule | Name: G4 RNA-mediated dimer of polycomb repressive complex 2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Polycomb protein SUZ12
| Macromolecule | Name: Polycomb protein SUZ12 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAPQKHGGGG GGGSGPSAGS GGGGFGGSAA VAAATASGGK SGGGSCGGGG SYSASSSSSA AAAAGAAVLP VKKPKMEHVQ ADHELFLQA FEKPTQIYRF LRTRNLIAPI FLHRTLTYMS HRNSRTNIKR KTFKVDDMLS KVEKMKGEQE SHSLSAHLQL T FTGFFHKN ...String: MAPQKHGGGG GGGSGPSAGS GGGGFGGSAA VAAATASGGK SGGGSCGGGG SYSASSSSSA AAAAGAAVLP VKKPKMEHVQ ADHELFLQA FEKPTQIYRF LRTRNLIAPI FLHRTLTYMS HRNSRTNIKR KTFKVDDMLS KVEKMKGEQE SHSLSAHLQL T FTGFFHKN DKPSPNSENE QNSVTLEVLL VKVCHKKRKD VSCPIRQVPT GKKQVPLNPD LNQTKPGNFP SLAVSSNEFE PS NSHMVKS YSLLFRVTRP GRREFNGMIN GETNENIDVN EELPARRKRN REDGEKTFVA QMTVFDKNRR LQLLDGEYEV AMQ EMEECP ISKKRATWET ILDGKRLPPF ETFSQGPTLQ FTLRWTGETN DKSTAPIAKP LATRNSESLH QENKPGSVKP TQTI AVKES LTTDLQTRKE KDTPNENRQK LRIFYQFLYN NNTRQQTEAR DDLHCPWCTL NCRKLYSLLK HLKLCHSRFI FNYVY HPKG ARIDVSINEC YDGSYAGNPQ DIHRQPGFAF SRNGPVKRTP ITHILVCRPK RTKASMSEFL ESEDGEVEQQ RTYSSG HNR LYFHSDTCLP LRPQEMEVDS EDEKDPEWLR EKTITQIEEF SDVNEGEKEV MKLWNLHVMK HGFIADNQMN HACMLFV EN YGQKIIKKNL CRNFMLHLVS MHDFNLISIM SIDKAVTKLR EMQQKLEKGE SASPANEEIT EEQNGTANGF SEINSKEK A LETDSVSGVS KQSKKQKL |
-Macromolecule #2: Polycomb protein EED
| Macromolecule | Name: Polycomb protein EED / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSEREVSTAP AGTDMPAAKK QKLSSDENSN PDLSGDENDD AVSIESGTNT ERPDTPTNTP NAPGRKSWGK GKWKSKKCKY SFKCVNSLK EDHNQPLFGV QFNWHSKEGD PLVFATVGSN RVTLYECHSQ GEIRLLQSYV DADADENFYT CAWTYDSNTS H PLLAVAGS ...String: MSEREVSTAP AGTDMPAAKK QKLSSDENSN PDLSGDENDD AVSIESGTNT ERPDTPTNTP NAPGRKSWGK GKWKSKKCKY SFKCVNSLK EDHNQPLFGV QFNWHSKEGD PLVFATVGSN RVTLYECHSQ GEIRLLQSYV DADADENFYT CAWTYDSNTS H PLLAVAGS RGIIRIINPI TMQCIKHYVG HGNAINELKF HPRDPNLLLS VSKDHALRLW NIQTDTLVAI FGGVEGHRDE VL SADYDLL GEKIMSCGMD HSLKLWRINS KRMMNAIKES YDYNPNKTNR PFISQKIHFP DFSTRDIHRN YVDCVRWLGD LIL SKSCEN AIVCWKPGKM EDDIDKIKPS ESNVTILGRF DYSQCDIWYM RFSMDFWQKM LALGNQVGKL YVWDLEVEDP HKAK CTTLT HHKCGAAIRQ TSFSRDSSIL IAVCDDASIW RWDRLR |
-Macromolecule #3: Histone-binding protein RBBP4
| Macromolecule | Name: Histone-binding protein RBBP4 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADKEAAFDD AVEERVINEE YKIWKKNTPF LYDLVMTHAL EWPSLTAQWL PDVTRPEGKD FSIHRLVLGT HTSDEQNHLV IASVQLPND DAQFDASHYD SEKGEFGGFG SVSGKIEIEI KINHEGEVNR ARYMPQNPCI IATKTPSSDV LVFDYTKHPS K PDPSGECN ...String: MADKEAAFDD AVEERVINEE YKIWKKNTPF LYDLVMTHAL EWPSLTAQWL PDVTRPEGKD FSIHRLVLGT HTSDEQNHLV IASVQLPND DAQFDASHYD SEKGEFGGFG SVSGKIEIEI KINHEGEVNR ARYMPQNPCI IATKTPSSDV LVFDYTKHPS K PDPSGECN PDLRLRGHQK EGYGLSWNPN LSGHLLSASD DHTICLWDIS AVPKEGKVVD AKTIFTGHTA VVEDVSWHLL HE SLFGSVA DDQKLMIWDT RSNNTSKPSH SVDAHTAEVN CLSFNPYSEF ILATGSADKT VALWDLRNLK LKLHSFESHK DEI FQVQWS PHNETILASS GTDRRLNVWD LSKIGEEQSP EDAEDGPPEL LFIHGGHTAK ISDFSWNPNE PWVICSVSED NIMQ VWQMA ENIYNDEDPE GSVDPEGQGS |
-Macromolecule #4: Histone-lysine N-methyltransferase EZH2
| Macromolecule | Name: Histone-lysine N-methyltransferase EZH2 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGQTGKKSEK GPVCWRKRVK SEYMRLRQLK RFRRADEVKS MFSSNRQKIL ERTEILNQEW KQRRIQPVHI LTSVSSLRGT RECSVTSDL DFPTQVIPLK TLNAVASVPI MYSWSPLQQN FMVEDETVLH NIPYMGDEVL DQDGTFIEEL IKNYDGKVHG D RECGFIND ...String: MGQTGKKSEK GPVCWRKRVK SEYMRLRQLK RFRRADEVKS MFSSNRQKIL ERTEILNQEW KQRRIQPVHI LTSVSSLRGT RECSVTSDL DFPTQVIPLK TLNAVASVPI MYSWSPLQQN FMVEDETVLH NIPYMGDEVL DQDGTFIEEL IKNYDGKVHG D RECGFIND EIFVELVNAL GQYNDDDDDD DGDDPEEREE KQKDLEDHRD DKESRPPRKF PSDKIFEAIS SMFPDKGTAE EL KEKYKEL TEQQLPGALP PECTPNIDGP NAKSVQREQS LHSFHTLFCR RCFKYDCFLH RKCNYSFHAT PNTYKRKNTE TAL DNKPCG PQCYQHLEGA KEFAAALTAE RIKTPPKRPG GRRRGRLPNN SSRPSTPTIN VLESKDTDSD REAGTETGGE NNDK EEEEK KDETSSSSEA NSRCQTPIKM KPNIEPPENV EWSGAEASMF RVLIGTYYDN FCAIARLIGT KTCRQVYEFR VKESS IIAP APAEDVDTPP RKKKRKHRLW AAHCRKIQLK KDGSSNHVYN YQPCDHPRQP CDSSCPCVIA QNFCEKFCQC SSECQN RFP GCRCKAQCNT KQCPCYLAVR ECDPDLCLTC GAADHWDSKN VSCKNCSIQR GSKKHLLLAP SDVAGWGIFI KDPVQKN EF ISEYCGEIIS QDEADRRGKV YDKYMCSFLF NLNNDFVVDA TRKGNKIRFA NHSVNPNCYA KVMMVNGDHR IGIFAKRA I QTGEELFFDY RYSQADALKY VGIEREMEIP |
-Macromolecule #5: protein Jumonji isoform X3
| Macromolecule | Name: protein Jumonji isoform X3 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSKERPKRNI IQKKYDDSDG IPWSEERVVR KVLYLSLKEF KNAQKRQHGE GIAGSLKSVN GLLGNDQSKA LGPASEQSEN EKDDASQVS STSNDVSSSD FEEGPSRKRP RLQAQRKFAQ SQPNSPSTTP VKTVEPLLPP PATQISDLSK RKPKTEDFLT F LCLRGSPA ...String: MSKERPKRNI IQKKYDDSDG IPWSEERVVR KVLYLSLKEF KNAQKRQHGE GIAGSLKSVN GLLGNDQSKA LGPASEQSEN EKDDASQVS STSNDVSSSD FEEGPSRKRP RLQAQRKFAQ SQPNSPSTTP VKTVEPLLPP PATQISDLSK RKPKTEDFLT F LCLRGSPA LPSSMVYFGS SQDEEDVEEE DDETEDVKTA NNNASSSCQS TPRKGKTHKH VHNGHVFNGS NRSTREKEPA QK HKSKETT PAKEKHIDHR ADSRREPASV AQPTATPSAG SLAKGLPANH QPPPPHRSAQ DLRKQVTLHV SKVNGVTRMS SLG AGTTSA KKIREVRPSP SKTVKYTATV TKGTVTYTKA KRELVKETKP THHKPSSAVN HTISGKTESS NAKTRKQVLS LGGA STSTG PAASGLKASS RLNPKSCTKE VGGRQLREGL RNSKRRLEEA QQVDKPQSPP KKMKGAAGIA EAPGKKASAA SAEKS LLNG HVKKEVPERS LERNRPKRAT AGKNMPGKQA HGKAEGTPCE NRSTSQPESS HKPHDPQGKP EKGIGKSGWT AMDEIP VLR PSAKEFHDPL IYIESVRAQV EKYGMCRVIP PPDWRPECKL NDEMRFVTQI QHIHKLGRRW GPNVQRLACI KKHLRSQ GI TMDELPLIGG CELDLACFFR LINEMGGMQQ VTDLKKWNKL ADMLRIPKTA QDRLAKLQEA YCQYLLSYDS LSPEEHRR L EKEVLMEKEI LEKRKGPLEG HTENDHHKFH SLPRFEPKNG LIHGVTPRNG FRSKLKEVGQ APLKTGRRRL FAQEKEVVK EEEEDKGVLN DFHKCIYKGR SVSLTTFYRT ARNIMNMCFS KEPAPAEIEQ EYWRLVEEKD CHVAVHCGKV DTNTHGSGFP VGKSEPFSR HGWNLTVLPN NTGSILRHLG AVPGVTIPWL NIGMVFSTSC WSRDQNHLPY IDYLHTGADC IWYCIPAEEE N KLEDVVHT LLQANGTPGL QMLESNVMIS PEVLCKEGIK VHRTVQQSGQ FVVCFPGSFV SKVCCGYSVS ETVHFATTQW TS MGFETAK EMKRRHIAKP FSMEKLLYQI AQAEAKKENG PTLSTISALL DELRDTELRQ RRQLFEAGLH SSARYGSHDG NST VADGKK KPRKWLQLET SERRCQICQH LCYLSMVVQE NENVVFCLEC ALRHVEKQKS CRGLKLMYRY DEEQIISLVN QICG KVSGK HGGIENCLNK PTPKRGPRKR ATVDVPPSRL PSS |
-Macromolecule #6: Zinc finger protein AEBP2
| Macromolecule | Name: Zinc finger protein AEBP2 / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAAITDMAD LEELSRLSPL PPGSPGSAAR GRAEPPEEEE EEEEEEEEAE AEAVAALLLN GGSGGGGGGG GGGVGGGEAE TMSEPSPES ASQAGEDEDE EEDDEEEEDE SSSSGGGEEE SSAESLVGSS GGSSSDETRS LSPGAASSSS GDGDGKEGLE E PKGPRGSQ ...String: MAAAITDMAD LEELSRLSPL PPGSPGSAAR GRAEPPEEEE EEEEEEEEAE AEAVAALLLN GGSGGGGGGG GGGVGGGEAE TMSEPSPES ASQAGEDEDE EEDDEEEEDE SSSSGGGEEE SSAESLVGSS GGSSSDETRS LSPGAASSSS GDGDGKEGLE E PKGPRGSQ GGGGGGSSSS SVVSSGGDEG YGTGGGGSSA TSGGRRGSLE MSSDGEPLSR MDSEDSISST IMDVDSTISS GR STPAMMN GQGSTTSSSK NIAYNCCWDQ CQACFNSSPD LADHIRSIHV DGQRGGVFVC LWKGCKVYNT PSTSQSWLQR HML THSGDK PFKCVVGGCN ASFASQGGLA RHVPTHFSQQ NSSKVSSQPK AKEESPSKAG MNKRRKLKNK RRRSLPRPHD FFDA QTLDA IRHRAICFNL SAHIESLGKG HSVVFHSTVI AKRKEDSGKI KLLLHWMPED ILPDVWVNES ERHQLKTKVV HLSKL PKDT ALLLDPNIYR TMPQKRLKRT LIRKVFNLYL SKQ |
-Macromolecule #7: G4 RNA
| Macromolecule | Name: G4 RNA / type: rna / ID: 7 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGGUAAGGGU AAGGGUAAGG GUAA |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 Details: RNP complex buffer (25 mM HEPES pH 7.9, 50 mM KCl, 2 mM MgCl2, 10% glycerol, and 1mM TCEP) EM preparation buffer I (25 mM HEPES pH 7.9, 50 mM KCl, 2.5% glycerol, and 1mM TCEP) EM preparation ...Details: RNP complex buffer (25 mM HEPES pH 7.9, 50 mM KCl, 2 mM MgCl2, 10% glycerol, and 1mM TCEP) EM preparation buffer I (25 mM HEPES pH 7.9, 50 mM KCl, 2.5% glycerol, and 1mM TCEP) EM preparation buffer II (25 mM HEPES pH 7.9, 50 mM KCl, 2.5% glycerol, 0.01%NP-40, and 1mM TCEP). |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 281 K / Instrument: LEICA PLUNGER / Details: 3s of single side blotting. |
| Details | We used streptavidin-affinity grid preparation method with biotin-labeled RNA at 100 nM concentration. PRC2 was applied in excess at 600 nM. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 0.01 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm Bright-field microscopy / Cs: 0.01 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Specialist optics | Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 18632 / Average electron dose: 60.0 e/Å2 / Details: 60 frames per movie |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X