+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of yeast Arginyltransferase 1 (ATE1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Arginyltransferase / post-translational modification / enzyme / TRANSFERASE-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationarginyltransferase / arginyl-tRNA--protein transferase activity / ubiquitin-dependent protein catabolic process via the N-end rule pathway / proteasomal protein catabolic process / apoptotic process / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Huang W / Zhang Y / Taylor DJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The structural basis of tRNA recognition by arginyl-tRNA-protein transferase. Authors: Thilini Abeywansha / Wei Huang / Xuan Ye / Allison Nawrocki / Xin Lan / Eckhard Jankowsky / Derek J Taylor / Yi Zhang /  Abstract: Arginyl-tRNA-protein transferase 1 (ATE1) is a master regulator of protein homeostasis, stress response, cytoskeleton maintenance, and cell migration. The diverse functions of ATE1 arise from its ...Arginyl-tRNA-protein transferase 1 (ATE1) is a master regulator of protein homeostasis, stress response, cytoskeleton maintenance, and cell migration. The diverse functions of ATE1 arise from its unique enzymatic activity to covalently attach an arginine onto its protein substrates in a tRNA-dependent manner. However, how ATE1 (and other aminoacyl-tRNA transferases) hijacks tRNA from the highly efficient ribosomal protein synthesis pathways and catalyzes the arginylation reaction remains a mystery. Here, we describe the three-dimensional structures of Saccharomyces cerevisiae ATE1 with and without its tRNA cofactor. Importantly, the putative substrate binding domain of ATE1 adopts a previously uncharacterized fold that contains an atypical zinc-binding site critical for ATE1 stability and function. The unique recognition of tRNA by ATE1 is coordinated through interactions with the major groove of the acceptor arm of tRNA. Binding of tRNA induces conformational changes in ATE1 that helps explain the mechanism of substrate arginylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29638.map.gz emd_29638.map.gz | 58.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29638-v30.xml emd-29638-v30.xml emd-29638.xml emd-29638.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29638.png emd_29638.png | 56.9 KB | ||
| Filedesc metadata |  emd-29638.cif.gz emd-29638.cif.gz | 6.4 KB | ||
| Others |  emd_29638_additional_1.map.gz emd_29638_additional_1.map.gz emd_29638_half_map_1.map.gz emd_29638_half_map_1.map.gz emd_29638_half_map_2.map.gz emd_29638_half_map_2.map.gz | 53.5 MB 2.3 MB 2.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29638 http://ftp.pdbj.org/pub/emdb/structures/EMD-29638 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29638 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29638 | HTTPS FTP |
-Related structure data
| Related structure data |  8fzrMC  8e3sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29638.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29638.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8726 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Phenix auto-sharpen map
| File | emd_29638_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phenix auto-sharpen map | ||||||||||||
| Projections & Slices |
| ||||||||||||
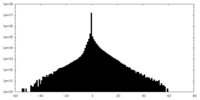
| Density Histograms |
-Half map: #2
| File | emd_29638_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
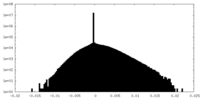
| Density Histograms |
-Half map: #1
| File | emd_29638_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : yeast Arginyltransferase 1 bound to Arg tRNA
| Entire | Name: yeast Arginyltransferase 1 bound to Arg tRNA |
|---|---|
| Components |
|
-Supramolecule #1: yeast Arginyltransferase 1 bound to Arg tRNA
| Supramolecule | Name: yeast Arginyltransferase 1 bound to Arg tRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Arginyl-tRNA--protein transferase 1
| Macromolecule | Name: Arginyl-tRNA--protein transferase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: arginyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 58.001277 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDRFVIWAP SMHNEPAAKC GYCHGNKGGN MDQLFALDSW AHRYMNKMDV VKIENCTIGS FVEHMDVATY DRMCNMGFRR SGKFLYKVD PLRNCCRLYT IRTAPQELNM TKELKKCISR FATRITSEDY CPAAVASSDF VGKIVNAEMN SKTFYTRFEP A LYSEEKYH ...String: MSDRFVIWAP SMHNEPAAKC GYCHGNKGGN MDQLFALDSW AHRYMNKMDV VKIENCTIGS FVEHMDVATY DRMCNMGFRR SGKFLYKVD PLRNCCRLYT IRTAPQELNM TKELKKCISR FATRITSEDY CPAAVASSDF VGKIVNAEMN SKTFYTRFEP A LYSEEKYH LFVKYQEKVH QDYNNSPKSF KRFLCDTPFG PEAVLGTQES WEQLNNWQRM KPGEKLKHMG PVHECYYYEG KL IAITVSD ILPSGISSVY FIWDPDYSKW SLGKLSALRD LAIIQRTNLQ YYYLGYYIED CPKMNYKANY GAEVLDVCHS KYI PLKPIQ DMISRGKLFV IGEEETKVTK ELYLVDSETG RGEGFPTDNV VKYKNIAEEI YGVGGCAFKS ANESALELKE LYGI PYEEE DLDTIYHLKE HNGHAPNGIP NVVPGLLPLW ELLDIMQSGK ITDLEGRLFL FEIETEGIRP LINFYSEPPN VKKRI CDVI RLFGFETCMK AVILYSEQM UniProtKB: Arginyl-tRNA--protein transferase 1 |
-Macromolecule #2: Arg tRNA
| Macromolecule | Name: Arg tRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.789736 KDa |
| Sequence | String: GCGCCCUUAG CUCAGUUGGA UAGAGCAACG ACCUUCUAAG UCGUGGGCCG CAGGUUCGAA UCCUGCAGGG CGCGCCA GENBANK: GENBANK: CP026935.2 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.1 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 25.0 µm / Nominal defocus min: 5.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)