+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
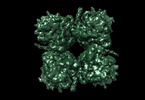
| Title | phiPA3 PhuN Tetramer, p2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Pseudomonas phage PhiPA3 (virus) Pseudomonas phage PhiPA3 (virus) | |||||||||
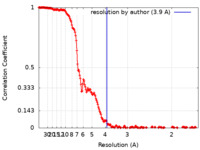
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Nieweglowska ES / Brilot AF / Mendez-Moran M / Kokontis C / Baek M / Li J / Cheng Y / Baker D / Bondy-Denomy J / Agard DA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The ϕPA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice. Authors: Eliza S Nieweglowska / Axel F Brilot / Melissa Méndez-Moran / Claire Kokontis / Minkyung Baek / Junrui Li / Yifan Cheng / David Baker / Joseph Bondy-Denomy / David A Agard /  Abstract: To protect themselves from host attack, numerous jumbo bacteriophages establish a phage nucleus-a micron-scale, proteinaceous structure encompassing the replicating phage DNA. Bacteriophage and host ...To protect themselves from host attack, numerous jumbo bacteriophages establish a phage nucleus-a micron-scale, proteinaceous structure encompassing the replicating phage DNA. Bacteriophage and host proteins associated with replication and transcription are concentrated inside the phage nucleus while other phage and host proteins are excluded, including CRISPR-Cas and restriction endonuclease host defense systems. Here, we show that nucleus fragments isolated from ϕPA3 infected Pseudomonas aeruginosa form a 2-dimensional lattice, having p2 or p4 symmetry. We further demonstrate that recombinantly purified primary Phage Nuclear Enclosure (PhuN) protein spontaneously assembles into similar 2D sheets with p2 and p4 symmetry. We resolve the dominant p2 symmetric state to 3.9 Å by cryo-EM. Our structure reveals a two-domain core, organized into quasi-symmetric tetramers. Flexible loops and termini mediate adaptable inter-tetramer contacts that drive subunit assembly into a lattice and enable the adoption of different symmetric states. While the interfaces between subunits are mostly well packed, two are open, forming channels that likely have functional implications for the transport of proteins, mRNA, and small molecules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29550.map.gz emd_29550.map.gz | 473.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29550-v30.xml emd-29550-v30.xml emd-29550.xml emd-29550.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29550_fsc.xml emd_29550_fsc.xml | 23.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29550.png emd_29550.png | 89.2 KB | ||
| Masks |  emd_29550_msk_1.map emd_29550_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_29550_half_map_1.map.gz emd_29550_half_map_1.map.gz emd_29550_half_map_2.map.gz emd_29550_half_map_2.map.gz | 23 MB 23 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29550 http://ftp.pdbj.org/pub/emdb/structures/EMD-29550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29550 | HTTPS FTP |
-Validation report
| Summary document |  emd_29550_validation.pdf.gz emd_29550_validation.pdf.gz | 672.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29550_full_validation.pdf.gz emd_29550_full_validation.pdf.gz | 671.7 KB | Display | |
| Data in XML |  emd_29550_validation.xml.gz emd_29550_validation.xml.gz | 26.2 KB | Display | |
| Data in CIF |  emd_29550_validation.cif.gz emd_29550_validation.cif.gz | 34.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29550 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29550 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29550 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29550 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29550.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29550.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29550_msk_1.map emd_29550_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29550_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29550_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Core tetramer assembly (p2) of the phiPA3 bacteriophage PhuN protein
| Entire | Name: Core tetramer assembly (p2) of the phiPA3 bacteriophage PhuN protein |
|---|---|
| Components |
|
-Supramolecule #1: Core tetramer assembly (p2) of the phiPA3 bacteriophage PhuN protein
| Supramolecule | Name: Core tetramer assembly (p2) of the phiPA3 bacteriophage PhuN protein type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage PhiPA3 (virus) Pseudomonas phage PhiPA3 (virus) |
-Macromolecule #1: Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN
| Macromolecule | Name: Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage PhiPA3 (virus) Pseudomonas phage PhiPA3 (virus) |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHMKIE EGKLVIWING DKGYNGLAEV GKKFEKDTGI KVTVEHPDKL EEKFPQVAAT GDGPDIIFWA HDRFGGYAQS GLLAEITPD KAFQDKLYPF TWDAVRYNGK LIAYPIAVEA LSLIYNKDLL PNPPKTWEEI PALDKELKAK GKSALMFNLQ E PYFTWPLI ...String: HHHHHHMKIE EGKLVIWING DKGYNGLAEV GKKFEKDTGI KVTVEHPDKL EEKFPQVAAT GDGPDIIFWA HDRFGGYAQS GLLAEITPD KAFQDKLYPF TWDAVRYNGK LIAYPIAVEA LSLIYNKDLL PNPPKTWEEI PALDKELKAK GKSALMFNLQ E PYFTWPLI AADGGYAFKY ENGKYDIKDV GVDNAGAKAG LTFLVDLIKN KHMNADTDYS IAEAAFNKGE TAMTINGPWA WS NIDTSKV NYGVTVLPTF KGQPSKPFVG VLSAGINAAS PNKELAKEFL ENYLLTDEGL EAVNKDKPLG AVALKSYEEE LAK DPRIAA TMENAQKGEI MPNIPQMSAF WYAVRTAVIN AASGRQTVDE ALKDAQTGKP IPNPLLGLDS TENLYFQGMQ QTQQ GPKVQ TQTLQGGAGN LNSIFQRSGR TDGGDARASE ALAVFNKLKE EAIAQQDLHD DFLVFRFDRD QNRVGYSALL VVKRA AING QQVIVTRPLV MPNDQITLPT KKLTIQNGMH QETIEAEADV QDVFTTQYWN RICDSIRQQT GKHDAMVINA GPTVIP ADF DLKDELVLKQ LLIKSVNLCD DMLAKRSGEQ PFSVAMLKGT DETLAARLNF TGKPMHDSLG YPIRSDILVS LNRVKKP GQ QENEFYEAED KLNQVSCFVN LEYTPQPQQA IYGAPQQTQQ LPPLTPAIVI TDVRQAEWLK ANTMELYLFA LSNAFRVT A NQSWARSLLP QLGKVKDMRD IGAIGYLSRL AARVETKTET FTDQNFAELL YNMVRPSPVF MSDLNRFGDN AAIENVFID ALGGVNQQRA VAAIIAGVNN LIGGGFEKFF DHNTMPIIQP YGTDIQLGYY LDGEGEKQDR RDLDVLGALN ASDGNIQEWM SWYGTQCNV AVHPELRARQ SKNFDRQYLG NSVTYTTRAH RGIWNPKFIE ALDKAIASVG LTVAMDNVAQ VFGAQRFSGN L AIADYAVT GTAQVSSGLV SNGGYNPQFG VGQGSGFY |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 Component:
Details: 0.25 cOmplete Protease Inhibitor Tablet also included | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)