+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Stanieria sp. CphA2 | |||||||||
 Map data Map data | Stanieria sp. CphA2 locally filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cyanophycin / CphA2 / ligase / ATP-grasp | |||||||||
| Function / homology |  Function and homology information Function and homology informationribosomal S6-glutamic acid ligase activity / SOS response / ATP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Stanieria sp. (bacteria) / Stanieria sp. (bacteria) /  Stanieria sp. NIES-3757 (bacteria) Stanieria sp. NIES-3757 (bacteria) | |||||||||
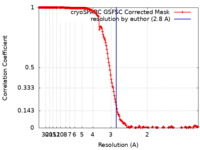
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Markus LM / Sharon I / Strauss M / Schmeing TM | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Protein Sci / Year: 2023 Journal: Protein Sci / Year: 2023Title: Structure and function of a hexameric cyanophycin synthetase 2. Authors: Linda M D Markus / Itai Sharon / Kim Munro / Marcel Grogg / Donald Hilvert / Mike Strauss / T Martin Schmeing /   Abstract: Cyanophycin is a natural polymer composed of a poly-aspartate backbone with arginine attached to each of the aspartate sidechains. Produced by a wide range of bacteria, which mainly use it as a store ...Cyanophycin is a natural polymer composed of a poly-aspartate backbone with arginine attached to each of the aspartate sidechains. Produced by a wide range of bacteria, which mainly use it as a store of fixed nitrogen, it has many promising industrial applications. Cyanophycin can be synthesized from the amino acids Asp and Arg by the widespread cyanophycin synthetase 1 (CphA1), or from the dipeptide β-Asp-Arg by the cyanobacterial enzyme cyanophycin synthetase 2 (CphA2). CphA2 enzymes display a range of oligomeric states, from dimers to dodecamers. Recently, the crystal structure of a CphA2 dimer was solved but could not be obtained in complex with substrate. Here, we report cryo-EM structures of the hexameric CphA2 from Stanieria sp. at ~2.8 Å resolution, both with and without ATP analog and cyanophycin. The structures show a two-fold symmetrical, trimer-of-dimers hexameric architecture, and substrate-binding interactions that are similar to those of CphA1. Mutagenesis experiments demonstrate the importance of several conserved substrate-binding residues. We also find that a Q416A/R528G double mutation prevents hexamer formation and use this double mutant to show that hexamerization augments the rate of cyanophycin synthesis. Together, these results increase our mechanistic understanding of how an interesting green polymer is biosynthesized. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29533.map.gz emd_29533.map.gz | 22.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29533-v30.xml emd-29533-v30.xml emd-29533.xml emd-29533.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29533_fsc.xml emd_29533_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_29533.png emd_29533.png | 64 KB | ||
| Masks |  emd_29533_msk_1.map emd_29533_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29533.cif.gz emd-29533.cif.gz | 5.5 KB | ||
| Others |  emd_29533_half_map_1.map.gz emd_29533_half_map_1.map.gz emd_29533_half_map_2.map.gz emd_29533_half_map_2.map.gz | 474.2 MB 474.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29533 http://ftp.pdbj.org/pub/emdb/structures/EMD-29533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29533 | HTTPS FTP |
-Related structure data
| Related structure data |  8fxhMC  8fxiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29533.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29533.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Stanieria sp. CphA2 locally filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.675 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29533_msk_1.map emd_29533_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Stanieria sp. CphA2 half map A
| File | emd_29533_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Stanieria sp. CphA2 half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Stanieria sp. CphA2 half map B
| File | emd_29533_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Stanieria sp. CphA2 half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Stanieria sp. CphA2
| Entire | Name: Stanieria sp. CphA2 |
|---|---|
| Components |
|
-Supramolecule #1: Stanieria sp. CphA2
| Supramolecule | Name: Stanieria sp. CphA2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Stanieria sp. (bacteria) Stanieria sp. (bacteria) |
-Macromolecule #1: RimK domain-containing protein ATP-grasp
| Macromolecule | Name: RimK domain-containing protein ATP-grasp / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Stanieria sp. NIES-3757 (bacteria) Stanieria sp. NIES-3757 (bacteria) |
| Molecular weight | Theoretical: 72.886516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLTKQAVEPV RINARTTDVF DIFNVKQYVG ANPYLNQAAL VFDFAFTESY QPLPIENYLA VVGDRYPRLK EIEYQSYAEL FASTVAEVN KLEMDLHLKG WNVKPIEEIN RIAIESLHHR TTKEVVYCVW DWFEFITQGE EFDLSKQIAI LQQLFRNSVY G GPTVYALL ...String: MLTKQAVEPV RINARTTDVF DIFNVKQYVG ANPYLNQAAL VFDFAFTESY QPLPIENYLA VVGDRYPRLK EIEYQSYAEL FASTVAEVN KLEMDLHLKG WNVKPIEEIN RIAIESLHHR TTKEVVYCVW DWFEFITQGE EFDLSKQIAI LQQLFRNSVY G GPTVYALL RTANEKHIPA FYLWDEGLMQ YGYGKQQVRG IATTFDVDSH IDSDFTTQKD DCKKFLQELG FPVPQGDVVF SL AEAKEVA AEIGYPVAVK PVAGHKGIGV TADVQDEIEL EAAYDRAVAG IPLEEKICII VENSIAGHDY RLLCVNGRFV AAT ERKPAY VVGDGYSTIA ELIEKENFSP NRSDTPTSPM GKIRTDEAMH LYLEEQGLDL DSVIDRDRTI YLRKVANLSS GGFS IDATN RVHPDNIILA QDIAQHFRLT CLGIDIITND IGRSWKETSF GIIEINAAPG VYMHLKPAIG EPVDVTARIL ETFFE TEKN ARIPIITFNR VSIRQLQKLS DRILMSHPDW TIGAVCREGI LINRSEKILN RHYNTNVLNL LRNPKLDLLI AEYDED ALE AEGMFYHGSN LVVLEDPSEI EMILTRDVFS DSTVIIKQGR EITIKRKGLL EQYELEAEEL IEQVYLKEIG TISENLY FQ UniProtKB: RimK domain-containing protein ATP-grasp |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: -0.0025 µm / Nominal defocus min: -0.001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)