[English] 日本語
 Yorodumi
Yorodumi- EMDB-29419: Cryo-EM structure of engineered hepatitis C virus E1E2 ectodomain... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of engineered hepatitis C virus E1E2 ectodomain in complex with antibodies AR4A, HEPC74, and IGH520 | |||||||||
 Map data Map data | Primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HCV / E1E2 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell mitochondrial membrane / host cell lipid droplet / viral nucleocapsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host innate immune response / host cell endoplasmic reticulum membrane / ribonucleoprotein complex / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...host cell mitochondrial membrane / host cell lipid droplet / viral nucleocapsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host innate immune response / host cell endoplasmic reticulum membrane / ribonucleoprotein complex / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell nucleus / virion membrane / structural molecule activity / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Hepacivirus C / synthetic construct (others) / Hepacivirus C / synthetic construct (others) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Metcalf MC / Ofek G | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies. Authors: Matthew C Metcalf / Benjamin M Janus / Rui Yin / Ruixue Wang / Johnathan D Guest / Edwin Pozharski / Mansun Law / Roy A Mariuzza / Eric A Toth / Brian G Pierce / Thomas R Fuerst / Gilad Ofek /  Abstract: Hepatitis C virus (HCV) is a major global health burden as the leading causative agent of chronic liver disease and hepatocellular carcinoma. While the main antigenic target for HCV-neutralizing ...Hepatitis C virus (HCV) is a major global health burden as the leading causative agent of chronic liver disease and hepatocellular carcinoma. While the main antigenic target for HCV-neutralizing antibodies is the membrane-associated E1E2 surface glycoprotein, the development of effective vaccines has been hindered by complications in the biochemical preparation of soluble E1E2 ectodomains. Here, we present a cryo-EM structure of an engineered, secreted E1E2 ectodomain of genotype 1b in complex with neutralizing antibodies AR4A, HEPC74, and IGH520. Structural characterization of the E1 subunit and C-terminal regions of E2 reveal an overall architecture of E1E2 that concurs with that observed for non-engineered full-length E1E2. Analysis of the AR4A epitope within a region of E2 that bridges between the E2 core and E1 defines the structural basis for its broad neutralization. Our study presents the structure of an E1E2 complex liberated from membrane via a designed scaffold, one that maintains all essential structural features of native E1E2. The study advances the understanding of the E1E2 heterodimer structure, crucial for the rational design of secreted E1E2 antigens in vaccine development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29419.map.gz emd_29419.map.gz | 167.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29419-v30.xml emd-29419-v30.xml emd-29419.xml emd-29419.xml | 33.7 KB 33.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29419.png emd_29419.png | 46.3 KB | ||
| Filedesc metadata |  emd-29419.cif.gz emd-29419.cif.gz | 7.2 KB | ||
| Others |  emd_29419_additional_1.map.gz emd_29419_additional_1.map.gz emd_29419_additional_2.map.gz emd_29419_additional_2.map.gz emd_29419_additional_3.map.gz emd_29419_additional_3.map.gz emd_29419_additional_4.map.gz emd_29419_additional_4.map.gz emd_29419_additional_5.map.gz emd_29419_additional_5.map.gz emd_29419_additional_6.map.gz emd_29419_additional_6.map.gz emd_29419_half_map_1.map.gz emd_29419_half_map_1.map.gz emd_29419_half_map_2.map.gz emd_29419_half_map_2.map.gz | 164.9 MB 167.8 MB 164.9 MB 88.7 MB 164.8 MB 88.7 MB 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29419 http://ftp.pdbj.org/pub/emdb/structures/EMD-29419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29419 | HTTPS FTP |
-Validation report
| Summary document |  emd_29419_validation.pdf.gz emd_29419_validation.pdf.gz | 906.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29419_full_validation.pdf.gz emd_29419_full_validation.pdf.gz | 906.5 KB | Display | |
| Data in XML |  emd_29419_validation.xml.gz emd_29419_validation.xml.gz | 14.7 KB | Display | |
| Data in CIF |  emd_29419_validation.cif.gz emd_29419_validation.cif.gz | 17.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29419 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29419 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29419 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29419 | HTTPS FTP |
-Related structure data
| Related structure data |  8fsjMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29419.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29419.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
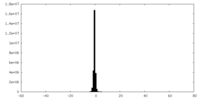
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Half map A for local refinement map
| File | emd_29419_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A for local refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
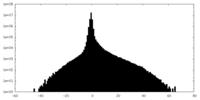
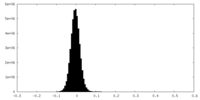
| Density Histograms |
-Additional map: Sharpened local refinement map
| File | emd_29419_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened local refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
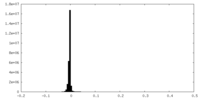
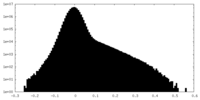
| Density Histograms |
-Additional map: Half map B for local refinement map
| File | emd_29419_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B for local refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
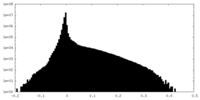
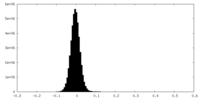
| Density Histograms |
-Additional map: Non-sharpened local refinement map
| File | emd_29419_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened local refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
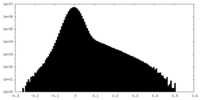
| Density Histograms |
-Additional map: Merged primary and local refinement map
| File | emd_29419_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Merged primary and local refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Non-sharpened primary map
| File | emd_29419_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened primary map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B for primary map
| File | emd_29419_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B for primary map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A for primary map
| File | emd_29419_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A for primary map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HCV E1E2 ectodomain in complex with AR4A, HEPC74, and IGH520
| Entire | Name: HCV E1E2 ectodomain in complex with AR4A, HEPC74, and IGH520 |
|---|---|
| Components |
|
-Supramolecule #1: HCV E1E2 ectodomain in complex with AR4A, HEPC74, and IGH520
| Supramolecule | Name: HCV E1E2 ectodomain in complex with AR4A, HEPC74, and IGH520 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Hepacivirus C Hepacivirus C |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: HCV E2 ectodomain, SYNZIP2 scaffold fusion
| Macromolecule | Name: HCV E2 ectodomain, SYNZIP2 scaffold fusion / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 42.975371 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STHVTGGTAS HTTRHFASLF SSGASQRVQL INTNGSWHIN RTALNCNDSL HTGFLAALFY THKFNASGCP ERMAHCRPID EFAQGWGPI TYAEGHGSDQ RPYCWHYAPR QCGTIPASQV CGPVYCFTPS PVVVGTTDRF GAPTYTWGEN ETDVLILNNT R PPQGNWFG ...String: STHVTGGTAS HTTRHFASLF SSGASQRVQL INTNGSWHIN RTALNCNDSL HTGFLAALFY THKFNASGCP ERMAHCRPID EFAQGWGPI TYAEGHGSDQ RPYCWHYAPR QCGTIPASQV CGPVYCFTPS PVVVGTTDRF GAPTYTWGEN ETDVLILNNT R PPQGNWFG CTWMNSTGFT KTCGGPPCNI GGVGNNTLTC PTDCFRKHPE ATYTKCGSGP WLTPRCLVDY PYRLWHYPCT VN FTIFKVR MYVGGVEHRL NAACNWTRGE RCDLQDRDRS ELSPLLLSTT EWQILPCSFT TLPALSTGLI HLHQNIVDVQ YLY GIGSAV VSFAIPGGLV AQLENEVASL ENENETLKKK NLHKKDLIAY LEKEIANLRK KIEHHHHHH UniProtKB: Genome polyprotein |
-Macromolecule #2: HEPC74 heavy chain
| Macromolecule | Name: HEPC74 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.644807 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGSSVKV SCTTSGGTYI NYAISWVRQA PGQGLEWVGG MSPISNTPKY AQKFQGRVTI TADESTSTTY MELSSLRPE DTAVYYCARD LLKYCGGGNC HSLLVDPWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW ...String: QVQLVQSGAE VKKPGSSVKV SCTTSGGTYI NYAISWVRQA PGQGLEWVGG MSPISNTPKY AQKFQGRVTI TADESTSTTY MELSSLRPE DTAVYYCARD LLKYCGGGNC HSLLVDPWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKRVEPK SCDKTAGWSH PQ FEKGGGS GGGSGGSSAW SHPQFEK |
-Macromolecule #3: HEPC74 light chain
| Macromolecule | Name: HEPC74 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.470111 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSPST LSASVGDRVT ISCRASQSIS SWLAWYQQKP GRAPKLLIYK ASSLETGVPS RFSGSGSGTE FTLTISSLQP DDFATYYCQ HYNTYLFTFG PGTKVDLKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIVMTQSPST LSASVGDRVT ISCRASQSIS SWLAWYQQKP GRAPKLLIYK ASSLETGVPS RFSGSGSGTE FTLTISSLQP DDFATYYCQ HYNTYLFTFG PGTKVDLKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #4: HCV E1 ectodomain, SYNZIP1 scaffold fusion
| Macromolecule | Name: HCV E1 ectodomain, SYNZIP1 scaffold fusion / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 22.951379 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YEVRNASGLY HVTNDCSNAS IVYETTDMIM HTPGCVPCVR EDNSSRCWVA LTPTLAARNA SVPTVAIRRH VDLLVGAAAF CSAMYVGDL CGSVFLVSQL FTFSPRRHET VQDCNCSIYP GHVSGHRMAW DMMMNWSPTA ALVVSQLLRI PQAVVDMVAP G GRNAYLRK ...String: YEVRNASGLY HVTNDCSNAS IVYETTDMIM HTPGCVPCVR EDNSSRCWVA LTPTLAARNA SVPTVAIRRH VDLLVGAAAF CSAMYVGDL CGSVFLVSQL FTFSPRRHET VQDCNCSIYP GHVSGHRMAW DMMMNWSPTA ALVVSQLLRI PQAVVDMVAP G GRNAYLRK KIARLKKDNL QLERDEQNLE KIIANLRDEI ARLENEVA UniProtKB: Genome polyprotein |
-Macromolecule #5: AR4A light chain
| Macromolecule | Name: AR4A light chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.743781 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGVPTQVLGL LLLWLTDARC EIELTLTQSP GTLSLSPGER ATLSCRASQS VSNNYLAWYQ QKPGQAPRLL IYGASSRATG IPDRFSGSG SGTGFTLIIS RLEPEDFAVY YCQQYGSSSI TFGQGTRLEI KRTVAAPSVF IFPPSDEQLK SGTASVVCLL N NFYREAKV ...String: MGVPTQVLGL LLLWLTDARC EIELTLTQSP GTLSLSPGER ATLSCRASQS VSNNYLAWYQ QKPGQAPRLL IYGASSRATG IPDRFSGSG SGTGFTLIIS RLEPEDFAVY YCQQYGSSSI TFGQGTRLEI KRTVAAPSVF IFPPSDEQLK SGTASVVCLL N NFYREAKV QWKVDNALQS GNSQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #6: AR4A heavy chain
| Macromolecule | Name: AR4A heavy chain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.862697 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYRMQLLSCI ALSLALVTNS EVQLLEQSGP EVKKPGDSLR ISCKMSGDSL VTTWIGWVRQ KPGQGLEWMG IINPGDSSTN IYPGDSATR YGPSFQGQVT ISIDKSTSTA YLQWNNVKAS DTGIYYCARH VPVPISGTFL WREREMHDFG YFDDWGQGTL V IVSSASTK ...String: MYRMQLLSCI ALSLALVTNS EVQLLEQSGP EVKKPGDSLR ISCKMSGDSL VTTWIGWVRQ KPGQGLEWMG IINPGDSSTN IYPGDSATR YGPSFQGQVT ISIDKSTSTA YLQWNNVKAS DTGIYYCARH VPVPISGTFL WREREMHDFG YFDDWGQGTL V IVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP EPVTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SL GTQTYIC NVNHKPSNTK VDKKVEPKSC AGWSHPQFEK GGGSGGGSGG SSAWSHPQFE K |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)