[English] 日本語
 Yorodumi
Yorodumi- EMDB-29347: Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex bound to a template DNA | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | DNA polymerase / primase / DNA replication / DNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information: / alpha DNA polymerase:primase complex / lagging strand elongation / mitotic DNA replication initiation / DNA replication, synthesis of primer / leading strand elongation / DNA replication origin binding / DNA replication initiation / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / single-stranded DNA binding ...: / alpha DNA polymerase:primase complex / lagging strand elongation / mitotic DNA replication initiation / DNA replication, synthesis of primer / leading strand elongation / DNA replication origin binding / DNA replication initiation / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / single-stranded DNA binding / 4 iron, 4 sulfur cluster binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / nucleotide binding / chromatin binding / DNA binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.6 Å | ||||||||||||
 Authors Authors | Yuan Z / Georgescu R / Li H / O'Donnell M | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation | Journal: bioRxiv / Year: 2023 Title: Molecular choreography of primer synthesis by the eukaryotic Pol α-primase. Authors: Zuanning Yuan / Roxana Georgescu / Huilin Li / Michael E O'Donnell /  Abstract: The eukaryotic polymerase α (Pol α) is a dual-function DNA polymerase/primase complex that synthesizes an RNA-DNA hybrid primer of 20-30 nucleotides for DNA replication. Pol α is composed of Pol1, ...The eukaryotic polymerase α (Pol α) is a dual-function DNA polymerase/primase complex that synthesizes an RNA-DNA hybrid primer of 20-30 nucleotides for DNA replication. Pol α is composed of Pol1, Pol12, Primase 1 (Pri1), and Pri2, with Pol1 and Pri1 containing the DNA polymerase activity and RNA primase activity, respectively, whereas Pol12 and Pri2 serve a structural role. It has been unclear how Pol α hands over an RNA primer made by Pri1 to Pol1 for DNA primer extension, and how the primer length is defined, perhaps due to the difficulty in studying the highly mobile structure. Here we report a comprehensive cryo-EM analysis of the intact 4-subunit yeast Pol α in the apo, primer initiation, primer elongation, RNA primer hand-off from Pri1 to Pol1, and DNA extension states in a 3.5 Å - 5.6 Å resolution range. We found that Pol α is a three-lobed flexible structure. Pri2 functions as a flexible hinge that holds together the catalytic Pol1-core, and the noncatalytic Pol1 CTD that binds to Pol 12 to form a stable platform upon which the other components are organized. In the apo state, Pol1-core is sequestered on the Pol12-Pol1-CTD platform, and Pri1 is mobile perhaps in search of a template. Upon binding a ssDNA template, a large conformation change is induced that enables Pri1 to perform RNA synthesis, and positions Pol1-core to accept the future RNA primed site 50 Å upstream of where Pri1 binds. We reveal in detail the critical point at which Pol1-core takes over the 3'-end of the RNA from Pri1. DNA primer extension appears limited by the spiral motion of Pol1-core while Pri2-CTD stably holds onto the 5' end of the RNA primer. Since both Pri1 and Pol1-core are attached via two linkers to the platform, primer growth will produce stress within this "two-point" attachment that may limit the length of the RNA-DNA hybrid primer. Hence, this study reveals the large and dynamic series of movements that Pol α undergoes to synthesize a primer for DNA replication. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29347.map.gz emd_29347.map.gz | 8.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29347-v30.xml emd-29347-v30.xml emd-29347.xml emd-29347.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29347.png emd_29347.png | 92.8 KB | ||
| Filedesc metadata |  emd-29347.cif.gz emd-29347.cif.gz | 7.6 KB | ||
| Others |  emd_29347_half_map_1.map.gz emd_29347_half_map_1.map.gz emd_29347_half_map_2.map.gz emd_29347_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29347 http://ftp.pdbj.org/pub/emdb/structures/EMD-29347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29347 | HTTPS FTP |
-Related structure data
| Related structure data |  8foeMC  8focC  8fodC  8fohC  8fojC  8fokC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29347.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29347.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
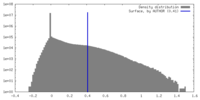
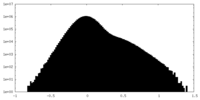
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29347_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
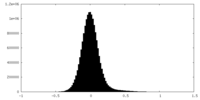
| Density Histograms |
-Half map: #1
| File | emd_29347_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
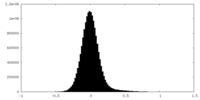
| Density Histograms |
- Sample components
Sample components
-Entire : DNA polymerase alpha/primase complex
| Entire | Name: DNA polymerase alpha/primase complex |
|---|---|
| Components |
|
-Supramolecule #1: DNA polymerase alpha/primase complex
| Supramolecule | Name: DNA polymerase alpha/primase complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2-#3, #1, #4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DNA polymerase
| Macromolecule | Name: DNA polymerase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 167.027766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSKSEKLEK LRKLQAARNG TSIDDYEGDE SDGDRIYDEI DEKEYRARKR QELLHDDFVV DDDGVGYVDR GVEEDWREVD NSSSDEDTG NLASKDSKRK KNIKREKDHQ ITDMLRTQHS KSTLLAHAKK SQKKSIPIDN FDDILGEFES GEVEKPNILL P SKLRENLN ...String: MSSKSEKLEK LRKLQAARNG TSIDDYEGDE SDGDRIYDEI DEKEYRARKR QELLHDDFVV DDDGVGYVDR GVEEDWREVD NSSSDEDTG NLASKDSKRK KNIKREKDHQ ITDMLRTQHS KSTLLAHAKK SQKKSIPIDN FDDILGEFES GEVEKPNILL P SKLRENLN SSPTSEFKSS IKRVNGNDES SHDAGISKKV KIDPDSSTDK YLEIESSPLK LQSRKLRYAN DVQDLLDDVE NS PVVATKR QNVLQDTLLA NPPSAQSLAD EEDDEDSDED IILKRRTMRS VTTTRRVNID SRSNPSTSPF VTAPGTPIGI KGL TPSKSL QSNTDVATLA VNVKKEDVVD PETDTFQMFW LDYCEVNNTL ILFGKVKLKD DNCVSAMVQI NGLCRELFFL PREG KTPTD IHEEIIPLLM DKYGLDNIRA KPQKMKYSFE LPDIPSESDY LKVLLPYQTP KSSRDTIPSD LSSDTFYHVF GGNSN IFES FVIQNRIMGP CWLDIKGADF NSIRNASHCA VEVSVDKPQN ITPTTTKTMP NLRCLSLSIQ TLMNPKENKQ EIVSIT LSA YRNISLDSPI PENIKPDDLC TLVRPPQSTS FPLGLAALAK QKLPGRVRLF NNEKAMLSCF CAMLKVEDPD VIIGHRL QN VYLDVLAHRM HDLNIPTFSS IGRRLRRTWP EKFGRGNSNM NHFFISDICS GRLICDIANE MGQSLTPKCQ SWDLSEMY Q VTCEKEHKPL DIDYQNPQYQ NDVNSMTMAL QENITNCMIS AEVSYRIQLL TLTKQLTNLA GNAWAQTLGG TRAGRNEYI LLHEFSRNGF IVPDKEGNRS RAQKQRQNEE NADAPVNSKK AKYQGGLVFE PEKGLHKNYV LVMDFNSLYP SIIQEFNICF TTVDRNKED IDELPSVPPS EVDQGVLPRL LANLVDRRRE VKKVMKTETD PHKRVQCDIR QQALKLTANS MYGCLGYVNS R FYAKPLAM LVTNKGREIL MNTRQLAESM NLLVVYGDTD SVMIDTGCDN YADAIKIGLG FKRLVNERYR LLEIDIDNVF KK LLLHAKK KYAALTVNLD KNGNGTTVLE VKGLDMKRRE FCPLSRDVSI HVLNTILSDK DPEEALQEVY DYLEDIRIKV ETN NIRIDK YKINMKLSKD PKAYPGGKNM PAVQVALRMR KAGRVVKAGS VITFVITKQD EIDNAADTPA LSVAERAHAL NEVM IKSNN LIPDPQYYLE KQIFAPVERL LERIDSFNVV RLSEALGLDS KKYFRREGGN NNGEDINNLQ PLETTITDVE RFKDT VTLE LSCPSCDKRF PFGGIVSSNY YRVSYNGLQC KHCEQLFTPL QLTSQIEHSI RAHISLYYAG WLQCDDSTCG IVTRQV SVF GKRCLNDGCT GVMRYKYSDK QLYNQLLYFD SLFDCEKNKK QELKPIYLPD DLDYPKEQLT ESSIKALTEQ NRELMET GR SVVQKYLNDC GRRYVDMTSI FDFMLN UniProtKB: DNA polymerase |
-Macromolecule #2: DNA primase
| Macromolecule | Name: DNA primase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.760367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTNSVKTNGP SSSDMEYYYK SLYPFKHIFN WLNHSPKPSR DMINREFAMA FRSGAYKRYN SFNSVQDFKA QIEKANPDRF EIGAIYNKP PRERDTLLKS ELKALEKELV FDIDMDDYDA FRTCCSGAQV CSKCWKFISL AMKITNTALR EDFGYKDFIW V FSGRRGAH ...String: MTNSVKTNGP SSSDMEYYYK SLYPFKHIFN WLNHSPKPSR DMINREFAMA FRSGAYKRYN SFNSVQDFKA QIEKANPDRF EIGAIYNKP PRERDTLLKS ELKALEKELV FDIDMDDYDA FRTCCSGAQV CSKCWKFISL AMKITNTALR EDFGYKDFIW V FSGRRGAH CWVSDKRARA LTDVQRRNVL DYVNVIRDRN TDKRLALKRP YHPHLARSLE QLKPFFVSIM LEEQNPWEDD QH AIQTLLP ALYDKQLIDS LKKYWLDNPR RSSKEKWNDI DQIATSLFKG PKQDSHIIKL RECKEDLVLM TLYPKLDVEV TKQ TIHLLK APFCIHPATG NVCVPIDESF APEKAPKLID LQTEMEKNND VSLTALQPFI NQFQAYVSSL LKNELGSVKR ERED DDEPA SLDF UniProtKB: DNA primase |
-Macromolecule #3: DNA primase large subunit
| Macromolecule | Name: DNA primase large subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.305457 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFRQSKRRIA SRKNFSSYDD IVKSELDVGN TNAANQIILS SSSSEEEKKL YARLYESKLS FYDLPPQGEI TLEQFEIWAI DRLKILLEI ESCLSRNKSI KEIETIIKPQ FQKLLPFNTE SLEDRKKDYY SHFILRLCFC RSKELREKFV RAETFLFKIR F NMLTSTDQ ...String: MFRQSKRRIA SRKNFSSYDD IVKSELDVGN TNAANQIILS SSSSEEEKKL YARLYESKLS FYDLPPQGEI TLEQFEIWAI DRLKILLEI ESCLSRNKSI KEIETIIKPQ FQKLLPFNTE SLEDRKKDYY SHFILRLCFC RSKELREKFV RAETFLFKIR F NMLTSTDQ TKFVQSLDLP LLQFISNEEK AELSHQLYQT VSASLQFQLN LNEEHQRKQY FQQEKFIKLP FENVIELVGN RL VFLKDGY AYLPQFQQLN LLSNEFASKL NQELIKTYQY LPRLNEDDRL LPILNHLSSG YTIADFNQQK ANQFSENVDD EIN AQSVWS EEISSNYPLC IKNLMEGLKK NHHLRYYGRQ QLSLFLKGIG LSADEALKFW SEAFTNNGNM TMEKFNKEYR YSFR HNYGL EGNRINYKPW DCHTILSKPR PGRGDYHGCP FRDWSHERLS AELRSMKLTQ AQIISVLDSC QKGEYTIACT KVFEM THNS ASADLEIGEQ THIAHPNLYF ERSRQLQKKQ QKLEKEKLFN NGNH UniProtKB: DNA primase large subunit |
-Macromolecule #4: DNA polymerase alpha subunit B
| Macromolecule | Name: DNA polymerase alpha subunit B / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.865938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGSIDVITH FGPDADKPEI ITALENLTKL HALSVEDLYI KWEQFSNQRR QTHTDLTSKN IDEFKQFLQL QMEKRANQIS SSSKVNTST KKPVIKKSLN SSPLFGLSIP KTPTLKKRKL HGPFSLSDSK QTYNVGSEAE TNEKGNSSLK LEFTPGMAED A VGDSAPLS ...String: MSGSIDVITH FGPDADKPEI ITALENLTKL HALSVEDLYI KWEQFSNQRR QTHTDLTSKN IDEFKQFLQL QMEKRANQIS SSSKVNTST KKPVIKKSLN SSPLFGLSIP KTPTLKKRKL HGPFSLSDSK QTYNVGSEAE TNEKGNSSLK LEFTPGMAED A VGDSAPLS HAKSSDAKTP GSSTFQTPTT NTPTTSRQNV PAGEILDSLN PENIEISSGN PNVGLLSTEE PSYNQVKVEP FY DAKKYKF RTMRQNLQEA SDVLDDQIES FTKIIQNHYK LSPNDFADPT IQSQSEIYAV GRIVPDSPTY DKFLNPESLS LET SRMGGV GRRVRLDLSQ VNELSFFLGQ IVAFKGKNAN GDYFTVNSIL PLPYPNSPVS TSQELQEFQA NLEGSSLKVI VTCG PYFAN DNFSLELLQE FIDSINNEVK PHVLIMFGPF IDITHPLIAS GKLPNFPQFK TQPKTLDELF LKLFTPILKT ISPHI QTVL IPSTKDAISN HAAYPQASLI RKALQLPKRN FKCMANPSSF QINEIYFGCS NVDTFKDLKE VIKGGTTSSR YRLDRV SEH ILQQRRYYPI FPGSIRTRIK PKDVSTKKET NDMESKEEKV YEHISGADLD VSYLGLTEFV GGFSPDIMII PSELQHF AR VVQNVVVINP GRFIRATGNR GSYAQITVQC PDLEDGKLTL VEGEEPVYLH NVWKRARVDL IAS UniProtKB: DNA polymerase alpha subunit B |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 176443 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)