[English] 日本語
 Yorodumi
Yorodumi- EMDB-29338: Cryo-EM structure of Cryptococcus neoformans trehalose-6-phosphat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Cryptococcus neoformans trehalose-6-phosphate synthase homotetramer in apo form | |||||||||
 Map data Map data | Cryptococcus neoformans trehalose-6-phosphate synthase (Tps1) homotetramer in the apo form | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glycosyltransferase / complex / transferase | |||||||||
| Function / homology |  Function and homology information Function and homology informationalpha,alpha-trehalose-phosphate synthase complex (UDP-forming) / alpha,alpha-trehalose-phosphate synthase (UDP-forming) / trehalose-phosphatase activity / alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity / trehalose biosynthetic process / cellular response to heat / cytosol Similarity search - Function | |||||||||
| Biological species |  Cryptococcus neoformans var. grubii H99 (fungus) Cryptococcus neoformans var. grubii H99 (fungus) | |||||||||
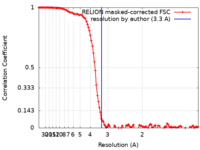
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Washington EJ / Brennan RG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Cryptococcus neoformans trehalose-6-phosphate synthase homotetramer in apo form Authors: Washington EJ / Brennan RG | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29338.map.gz emd_29338.map.gz | 117.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29338-v30.xml emd-29338-v30.xml emd-29338.xml emd-29338.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29338_fsc.xml emd_29338_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29338.png emd_29338.png | 111 KB | ||
| Filedesc metadata |  emd-29338.cif.gz emd-29338.cif.gz | 5.4 KB | ||
| Others |  emd_29338_half_map_1.map.gz emd_29338_half_map_1.map.gz emd_29338_half_map_2.map.gz emd_29338_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29338 http://ftp.pdbj.org/pub/emdb/structures/EMD-29338 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29338 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29338 | HTTPS FTP |
-Related structure data
| Related structure data |  8fo1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29338.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29338.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryptococcus neoformans trehalose-6-phosphate synthase (Tps1) homotetramer in the apo form | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryptococcus neoformans trehalose-6-phosphate synthase (Tps1) homotetramer in the...
| File | emd_29338_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryptococcus neoformans trehalose-6-phosphate synthase (Tps1) homotetramer in the apo half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
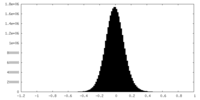
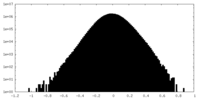
| Density Histograms |
-Half map: Cryptococcus neoformans trehalose-6-phosphate synthase (Tps1) homotetramer in the...
| File | emd_29338_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryptococcus neoformans trehalose-6-phosphate synthase (Tps1) homotetramer in the apo half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
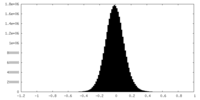
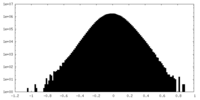
| Density Histograms |
- Sample components
Sample components
-Entire : trehalose-6-phosphate synthase homotetramer in apo form
| Entire | Name: trehalose-6-phosphate synthase homotetramer in apo form |
|---|---|
| Components |
|
-Supramolecule #1: trehalose-6-phosphate synthase homotetramer in apo form
| Supramolecule | Name: trehalose-6-phosphate synthase homotetramer in apo form type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Cryptococcus neoformans var. grubii H99 (fungus) Cryptococcus neoformans var. grubii H99 (fungus) |
| Molecular weight | Theoretical: 307 KDa |
-Macromolecule #1: Alpha,alpha-trehalose-phosphate synthase (UDP-forming)
| Macromolecule | Name: Alpha,alpha-trehalose-phosphate synthase (UDP-forming) type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cryptococcus neoformans var. grubii H99 (fungus) Cryptococcus neoformans var. grubii H99 (fungus) |
| Molecular weight | Theoretical: 76.8155 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHSSG VDLGTENLYF QSNAMTTMSN DIPNSPTSTS FTGTFSPAAT AANTAANART SDAPSPTTSS SGPKLETSKE QRLIVVSNR LPVTISKDDN GEYHFKMSSG GLVSALSGCK KTMSFTWIGW PGKDIPMQDR ETVNRRLLDE YNCYPVYLSD E LADSHYNG ...String: MHHHHHHSSG VDLGTENLYF QSNAMTTMSN DIPNSPTSTS FTGTFSPAAT AANTAANART SDAPSPTTSS SGPKLETSKE QRLIVVSNR LPVTISKDDN GEYHFKMSSG GLVSALSGCK KTMSFTWIGW PGKDIPMQDR ETVNRRLLDE YNCYPVYLSD E LADSHYNG FSNSILWPLF HYHPGEMNFD AAHWLAYREA NMRFADVVSS LVQAGDMVWV QDYHLMLLPM LLRSMITGES AQ GEMVRQE LGRVKEGVDD TVVKEVLKMG PGVAQAEDEG VEMLDDVEEE GGEMDVKSSP KRPHYARGMS TFQKQELVAK EKG KEGIRI GFFLHTPFPS SEIYRILPVR REILLGVLQC DLIGFHTYDY ARHFLSSCTR ILGLETQPNG IEFDGRYCQV GTFP IGIDP NQFIEGLQKE SIVKRLRSLE ARFEGVKVII GVDRLDYIKG IPQKLQALET FLTQHPEWIG KVVLVQLAIP SRQDV EEYQ DLRACVNELV GRINGRFGTV ESVPIHYMHK SVPFEELTAM YALADACLVT STRDGMNLVA YEYISSQAER HGSMIL SEF AGAAQSFNGS LLINPWDVQS TADAINQALT LSPQQRKTNW QKLFNYVSKY TAEAWGVSFV NELNRLSGQR PSGPTGL AG RRKSGSLSRT SSKASIQRRK SSQSGIVTGL GAAAGAAVNW AQAQVQGGSQ T UniProtKB: alpha,alpha-trehalose-phosphate synthase (UDP-forming) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)