[English] 日本語
 Yorodumi
Yorodumi- EMDB-29245: Cryo-EM structure of native mosquito salivary gland surface prote... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of native mosquito salivary gland surface protein 1 (SGS1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rhs/YD-repeats / mosquito salivary protein / receptor domain / pathogen transmission / ALLERGEN | |||||||||
| Function / homology | Tox-SGS domain / Salivary glad secreted protein domain toxin / Rhs repeat-associated core / : / membrane / AAEL009993-PA Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
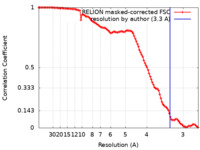
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Liu S / Xia X / Calvo E / Zhou ZH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Native structure of mosquito salivary protein uncovers domains relevant to pathogen transmission. Authors: Shiheng Liu / Xian Xia / Eric Calvo / Z Hong Zhou /  Abstract: Female mosquitoes inject saliva into vertebrate hosts during blood feeding. This process transmits mosquito-borne human pathogens that collectively cause ~1,000,000 deaths/year. Among the most ...Female mosquitoes inject saliva into vertebrate hosts during blood feeding. This process transmits mosquito-borne human pathogens that collectively cause ~1,000,000 deaths/year. Among the most abundant and conserved proteins secreted by female salivary glands is a high-molecular weight protein called salivary gland surface protein 1 (SGS1) that facilitates pathogen transmission, but its mechanism remains elusive. Here, we determine the native structure of SGS1 by the cryoID approach, showing that the 3364 amino-acid protein has a Tc toxin-like Rhs/YD shell, four receptor domains, and a set of C-terminal daisy-chained helices. These helices are partially shielded inside the Rhs/YD shell and poised to transform into predicted transmembrane helices. This transformation, and the numerous receptor domains on the surface of SGS1, are likely key in facilitating sporozoite/arbovirus invasion into the salivary glands and manipulating the host's immune response. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29245.map.gz emd_29245.map.gz | 96.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29245-v30.xml emd-29245-v30.xml emd-29245.xml emd-29245.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29245_fsc.xml emd_29245_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_29245.png emd_29245.png | 80.6 KB | ||
| Filedesc metadata |  emd-29245.cif.gz emd-29245.cif.gz | 8 KB | ||
| Others |  emd_29245_half_map_1.map.gz emd_29245_half_map_1.map.gz emd_29245_half_map_2.map.gz emd_29245_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29245 http://ftp.pdbj.org/pub/emdb/structures/EMD-29245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29245 | HTTPS FTP |
-Related structure data
| Related structure data |  8fjpMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29245.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29245.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29245_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
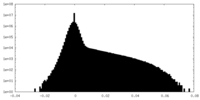
| Projections & Slices |
| ||||||||||||
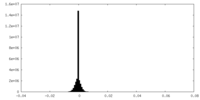
| Density Histograms |
-Half map: #1
| File | emd_29245_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
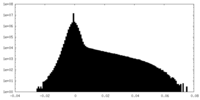
| Projections & Slices |
| ||||||||||||
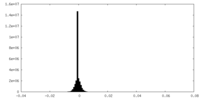
| Density Histograms |
- Sample components
Sample components
-Entire : salivary gland surface protein 1 (SGS1)
| Entire | Name: salivary gland surface protein 1 (SGS1) |
|---|---|
| Components |
|
-Supramolecule #1: salivary gland surface protein 1 (SGS1)
| Supramolecule | Name: salivary gland surface protein 1 (SGS1) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: salivary gland surface protein 1
| Macromolecule | Name: salivary gland surface protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 380.453688 KDa |
| Sequence | String: MLRIKEVKKY NNRLGFPAGK EVRIRPLLEA GGFARGPIYV KYGRELSKHD PSSRSMAWNA TVFDTALNHP TRPWTVTVVG GKKMVLGVR KEGLGFSEDD PPRSLNVKGG DDMADENYGG GNSTIVLVKS GGKDFAIGLD DLKELNSFGI GAGNGKLSLA L GNPVNVLT ...String: MLRIKEVKKY NNRLGFPAGK EVRIRPLLEA GGFARGPIYV KYGRELSKHD PSSRSMAWNA TVFDTALNHP TRPWTVTVVG GKKMVLGVR KEGLGFSEDD PPRSLNVKGG DDMADENYGG GNSTIVLVKS GGKDFAIGLD DLKELNSFGI GAGNGKLSLA L GNPVNVLT GVVLPVEQRD GSVAYVAVNS DTKSVIYKVD AKGLQKNQKI DVNVRASAEK ILLTKNNELL HISPQGLNVY SV QGSASTF KYFCPYFSSF GGWSRRFVDT VTLMDEGGNQ EALIGTGPKG IEYMPVDEKC KNYVVEGVQD AKHAVVAPGA TKE GNDIKV LGVYNGELCL LTLEVVDAVE PKLDQGSPAK PSAERVKATK SSKVTVRSGA LSKPVSWLRD SLDEASFKNI VDKL SGKIR FSFPLVDLQG RMGLPIKLVV YYDESDGEDM SVLGRHWTLG RDCIVLDHGN TVFEDKQDYY LVKDQMKIRL ERDWT KPSA NGKVLFNMVG NKDATFEYTA REEKWEVSDG KIRYVYGMNN QGVVSVPGWA DWYGPSANYD SKKIHSVQWN LVEISS VAH RDVKLKYTYT PLDPSTHTMH LLSITDDSNK TTIKFSYKTI EGLGKQVSSG LVKEHKFINN KLLEKVEIDS PSTAQVL KL TSTKIDSLYY LESIKQDDDP DPVLGFEYNK DDKLKPRVQQ IRLPSKSVVD FKYTKQAIAT QQFEQEIAKM ADLYTGNE Y SLEIEKTVGE TDLYVRLKDS AGKNDFIKNQ SIRIESYRGF KIKSYSPFMM HSYIAILVRY AEEKEKLHNK IYILNKGED DSWKLDTTNN NSRVSEDKKF KYDFQEDSFV YYHSKKVHFE YKKSGTKVWS YTSKDIGDVD AFTLMNRGAV YCKNDLVLIR WDALGKLQQ ETLSDAKSKP SISDVDTFFE YIDVQGTFPE DEAEAKKEVD DYKRDLKESL SDYGLILYNN VVALRTIKLS F TGRITVKV LLYLLKHDYT VSSRSSIELQ GGDLAKFNLT LDVFDEHKTK NTNDLDKYRF EFKKQGSKFK LTYIEATDKD NK PTKPSSQ NEKLIGQYER RMKIPLDFEK YMMQVNQEGI IVNDHQIIHD NGNFIAKQLD RDTLKLTKFK IPLGAFSNFK KDS DGDEIK LCTKTEQERT ESCVSLQTNS ARNVSIKYPY YLVTQKKNDI KVLPLKINSR GWEDSVDYRG EILHGSSSHA AMVT TRMSD QKTIVRPLKA LNKINKIYAQ VISEEKLTTP YDSYVIKYEY EDPVVSMNQV AFKTTIVVPG GGKSATGYYR ETNDL QDNQ QIVQVMTADN QVFDPEYVKR MNEMQQEEDK QRDGAQLDAE QTITDKSGYH PILKTTPYSA NQELVQFLGF EDYEDM TGW TVNKRPIQES NIRRNEFSAT GRNFLLLKKG EELIAEFPNT AYYDNFIAST WIRTAQEATV GSTTDMLLLY VDNKPMK GT IKQSIDEWIY VEADSREIVV SESTTVKRVL FKIIVKSTGA DDVHVDHVRL SPVNFNFEGS VYDARIGQRT ATIQTNGF V SRRLYDAYNR RIAEVDETGN IKYLASYSKR VGNTKDEKKR EGGYSSRIQM RAKHSWVESF SPYTMEKRWQ IGGTATVEP NQAILQGQII SKEKFSSESI CIRLVYSMSG SSQLSLTIGK TTVVVKPNAV QYKGHTATTP SNAELVIFAT PKLTSIWVDG HLRIEAPET HAKFNNEAVS LQTSGPVGIK DVIVMEDAEI QVSYLNRDSK PLQEILLLDS SNVLIRQMMY DVIGRRVAET V WVQKSLID GRSTAFKAFQ YHDDFVSNDN PTDRNYFLNT GPMQGYVATA TNTIYEGYPY SQTVYYNNPL EIRHKVGHPG VK NSIKGAF VHQYAIASDL AFIQRNYPKN EGYRQEEEKS PNGKKHVVVY NRRNKKVAEY TQVKDYNNIL TTYIYDQHGN QIQ MLPPSY YHEKSRSGDY QPEKQVVASP WAVTSKYDST GEFITSKETP DGGRVEFIYN EYNQLRYQIH YKEDKQADKI VYFL YNIFG RMCEAGQVPA NPTTLQQVRE TTKSHQAIPN RDQAVYFDYG ETESEPSLRG RIQRTVKKNK EVLFSEVMFF DEESN IIRK SYISPTTNET LSLVYLQEND KVSGIQYPFG VDGKQLILKY KHNLRGEIVE VARVEQKTSG QTEFIPIAGI DHDAEG KVT KISHNYGDSK FDQTYKYVAP GYLVEIANNF LTEKLYYTEK GYGCEPTGDG SILRTEFKAS WHDKCDQNLI PLTARAF VS GGIDFTTAET CFDALLNLGY IDTTGRPVKT FYPDLETGLP MKCATPSNWR YISEKMLEQG YPEHYGHAYD YGSHGELI A AKSFVGKEKD SLTAPLSKAS FANAGMKSHE LDRFWDSLSR SINKVEGTKA IFEGTQQLTT GLVGSVIHKP KLESLLEGK GGDSSICTPW SSGDRTEEAK CKREYQQAFD KLKLKQVIQS LQEPVRKNVL RILKNTLASM LGNSPGDVES FSIDPNGNHG VFYTGFKRF ELKYKHQKNQ IATIKEGTKQ QEKMIVHDDE GNVIKALHKK IDKIEYDPLT QRVSRIEMSD RSRTLEFGYD F RGERTFKR VRNKDNDIIS VNYYVRDNKG NVLVEYKQEY PNPKDTNKPI NTVTAYIHGP LGLLGFFRNN KYYNVLLDHE GS TRLVIHQ GKVVAAYDYL PYGQMIRKYG SNPEAHIAFR YTGQEFDEET GLYNYHARLY DPDIGRFFQM DPMEQYASPY KYA GNSPVS QIDPDGQIAV TLVLMIIGAI VGAYLGAASA NNSWNPAKWA WGDKKTWIGL FAGAIMGAFA VYGGAATFSY FTAM FGGSM IAGALATGVI SVAGAFLGAA AASNQWNPAK WDWTSPAVWN GLLSGASIAV SFPSGFVGIT RSFMSISSNL VKMIY ASLM VGGFLLFVYL GGGMANNFNF QISQWDWKSP RTWFGMIEGA STIFMGTAGT AKHGAAKVYN VVKPNGLKMI WHKVNI PSK AFTMRRVKDT IILTWYKNGQ SISKQILKTT VKADLAKIPK DFIMIHRGFF MPYQRIGYAA IAMPSMAGLM FKKNQYF FT NHPNGTLTKH VRKKRSAPMS SSAASPSVSN FLNDFFENMS ELFDSFFSQT EHSHQDSQSS LSIGASYGRP SNESYHKS F QKLCYSPDSD GNQIICPQRE STVNIFSKGE TFAPEAFGQD LFSRCLPLTW HDRPSIACDG EQTTFIYTPN QNIRVFDMV DGWLMLARIA PAALRNLKAG FSFLRDVVFS DEREQTVQVN DLSRCKQDLE VELLDLKRVM LKKQPNEVKW AQPILNDLED DIGEFLSER KPSEKEFELL QERLSALREE IMENSSVATE LNLSTLIGDM LKKMDGVNVG LNGDVRDMIS TLSGMVPFSS S NLLA UniProtKB: AAEL009993-PA |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4, pH 7.4 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Overall B value: 109.4 |
|---|---|
| Output model |  PDB-8fjp: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)