+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of a broadly neutralizing antibody STI-9167 | |||||||||
 Map data Map data | non-sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibody / SARS-CoV-2 / IMMUNE SYSTEM | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
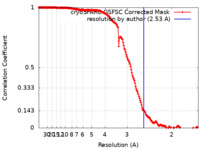
| Method | single particle reconstruction / cryo EM / Resolution: 2.53 Å | |||||||||
 Authors Authors | Bajic G | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2024 Journal: mBio / Year: 2024Title: An experimental pipeline to characterize the epitope of a SARS-CoV-2 neutralizing antibody. Authors: Kristina E Atanasoff / Luca Brambilla / Daniel C Adelsberg / Shreyas Kowdle / Christian S Stevens / Stefan Slamanig / Chuan-Tien Hung / Yanwen Fu / Reyna Lim / Linh Tran / Robert Allen / ...Authors: Kristina E Atanasoff / Luca Brambilla / Daniel C Adelsberg / Shreyas Kowdle / Christian S Stevens / Stefan Slamanig / Chuan-Tien Hung / Yanwen Fu / Reyna Lim / Linh Tran / Robert Allen / Weina Sun / J Andrew Duty / Goran Bajic / Benhur Lee / Domenico Tortorella /   Abstract: The COVID-19 pandemic remains a significant public health concern for the global population; the development and characterization of therapeutics, especially ones that are broadly effective, will ...The COVID-19 pandemic remains a significant public health concern for the global population; the development and characterization of therapeutics, especially ones that are broadly effective, will continue to be essential as severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) variants emerge. Neutralizing monoclonal antibodies remain an effective therapeutic strategy to prevent virus infection and spread so long as they recognize and interact with circulating variants. The epitope and binding specificity of a neutralizing anti-SARS-CoV-2 Spike receptor-binding domain antibody clone against many SARS-CoV-2 variants of concern were characterized by generating antibody-resistant virions coupled with cryo-EM structural analysis and VSV-spike neutralization studies. This workflow can serve to predict the efficacy of antibody therapeutics against emerging variants and inform the design of therapeutics and vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29227.map.gz emd_29227.map.gz | 253.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29227-v30.xml emd-29227-v30.xml emd-29227.xml emd-29227.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29227_fsc.xml emd_29227_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29227.png emd_29227.png | 94.6 KB | ||
| Filedesc metadata |  emd-29227.cif.gz emd-29227.cif.gz | 4.3 KB | ||
| Others |  emd_29227_additional_1.map.gz emd_29227_additional_1.map.gz emd_29227_additional_2.map.gz emd_29227_additional_2.map.gz emd_29227_half_map_1.map.gz emd_29227_half_map_1.map.gz emd_29227_half_map_2.map.gz emd_29227_half_map_2.map.gz | 451.4 MB 483.6 MB 475.9 MB 475.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29227 http://ftp.pdbj.org/pub/emdb/structures/EMD-29227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29227 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29227.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29227.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: DeepEMhancer sharpened map
| File | emd_29227_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
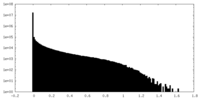
| Density Histograms |
-Additional map: cryoSPARC sharpened map
| File | emd_29227_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoSPARC sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
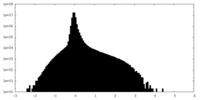
| Density Histograms |
-Half map: half-map 2
| File | emd_29227_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
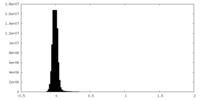
| Density Histograms |
-Half map: half-map 1
| File | emd_29227_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
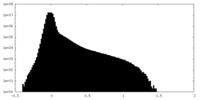
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike RBD in complex with 10A3 Fab
| Entire | Name: SARS-CoV-2 spike RBD in complex with 10A3 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike RBD in complex with 10A3 Fab
| Supramolecule | Name: SARS-CoV-2 spike RBD in complex with 10A3 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 58.92 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)