[English] 日本語
 Yorodumi
Yorodumi- EMDB-29042: Structure of iSAT large ribosomal subunit assembly intermediate -... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of iSAT large ribosomal subunit assembly intermediate - Class B2 | ||||||||||||
 Map data Map data | Class B2 iSAT 50S Ribosomal Intermediate | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | 50S subunit / assembly intermediate / in vitro / RIBOSOME | ||||||||||||
| Biological species |  | ||||||||||||
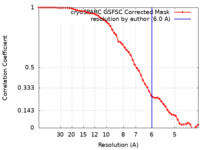
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | ||||||||||||
 Authors Authors | Dong X / Doerfel LK / Sheng K / Rabuck-Gibbons JN / Popova AM / Lyumkis D / Williamson JR | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Near-physiological in vitro assembly of 50S ribosomes involves parallel pathways. Authors: Xiyu Dong / Lili K Doerfel / Kai Sheng / Jessica N Rabuck-Gibbons / Anna M Popova / Dmitry Lyumkis / James R Williamson /  Abstract: Understanding the assembly principles of biological macromolecular complexes remains a significant challenge, due to the complexity of the systems and the difficulties in developing experimental ...Understanding the assembly principles of biological macromolecular complexes remains a significant challenge, due to the complexity of the systems and the difficulties in developing experimental approaches. As a ribonucleoprotein complex, the ribosome serves as a model system for the profiling of macromolecular complex assembly. In this work, we report an ensemble of large ribosomal subunit intermediate structures that accumulate during synthesis in a near-physiological and co-transcriptional in vitro reconstitution system. Thirteen pre-50S intermediate maps covering the entire assembly process were resolved using cryo-EM single-particle analysis and heterogeneous subclassification. Segmentation of the set of density maps reveals that the 50S ribosome intermediates assemble based on fourteen cooperative assembly blocks, including the smallest assembly core reported to date, which is composed of a 600-nucleotide-long folded rRNA and three ribosomal proteins. The cooperative blocks assemble onto the assembly core following defined dependencies, revealing the parallel pathways at both early and late assembly stages of the 50S subunit. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29042.map.gz emd_29042.map.gz | 8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29042-v30.xml emd-29042-v30.xml emd-29042.xml emd-29042.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29042_fsc.xml emd_29042_fsc.xml | 5.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_29042.png emd_29042.png | 79 KB | ||
| Filedesc metadata |  emd-29042.cif.gz emd-29042.cif.gz | 4.7 KB | ||
| Others |  emd_29042_half_map_1.map.gz emd_29042_half_map_1.map.gz emd_29042_half_map_2.map.gz emd_29042_half_map_2.map.gz | 14.5 MB 14.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29042 http://ftp.pdbj.org/pub/emdb/structures/EMD-29042 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29042 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29042 | HTTPS FTP |
-Validation report
| Summary document |  emd_29042_validation.pdf.gz emd_29042_validation.pdf.gz | 905.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29042_full_validation.pdf.gz emd_29042_full_validation.pdf.gz | 905.1 KB | Display | |
| Data in XML |  emd_29042_validation.xml.gz emd_29042_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_29042_validation.cif.gz emd_29042_validation.cif.gz | 15.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29042 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29042 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29042 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29042 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29042.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29042.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class B2 iSAT 50S Ribosomal Intermediate | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.096 Å | ||||||||||||||||||||||||||||||||||||
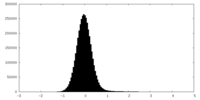
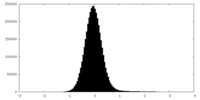
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Class B2 iSAT 50S Ribosomal Intermediate - Half Map 1
| File | emd_29042_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class B2 iSAT 50S Ribosomal Intermediate - Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
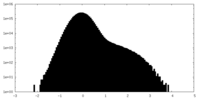
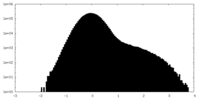
| Density Histograms |
-Half map: Class B2 iSAT 50S Ribosomal Intermediate - Half Map 2
| File | emd_29042_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class B2 iSAT 50S Ribosomal Intermediate - Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : iSAT large ribosomal subunit assembly intermediate (Class B2)
| Entire | Name: iSAT large ribosomal subunit assembly intermediate (Class B2) |
|---|---|
| Components |
|
-Supramolecule #1: iSAT large ribosomal subunit assembly intermediate (Class B2)
| Supramolecule | Name: iSAT large ribosomal subunit assembly intermediate (Class B2) type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: Most of the sucrose was removed by spin concentration. | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 / Details: 3 microliter of the sample was added.. | ||||||||||||||||||
| Details | The in vitro assembled large ribosomal subunit was purified by sucrose gradient and was spin-concentrated in a 100 kDa MW-cutoff filter. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | In order to account for highly preferred orientation of the specimen, data were acquired using tilts ranging from 10-60 degrees, in addition to untilted images at 0 degrees. The CTF was estimated on a per-particle basis to account for the gradient of CTF values across individual micrographs |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-50 / Number real images: 4607 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)