[English] 日本語
 Yorodumi
Yorodumi- EMDB-29028: CryoEM structure of Conalbumin from chicken egg white (sigma-Cas ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Conalbumin from chicken egg white (sigma-Cas 1391-06-6) | |||||||||
 Map data Map data | Final map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | conalbumin / egg white / ovotransferrin / iron complex / METAL TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationorganomineral extracellular matrix / iron ion transmembrane transport / antimicrobial humoral response / ferric iron binding / acute-phase response / iron ion transport / recycling endosome / antibacterial humoral response / response to lipopolysaccharide / intracellular iron ion homeostasis ...organomineral extracellular matrix / iron ion transmembrane transport / antimicrobial humoral response / ferric iron binding / acute-phase response / iron ion transport / recycling endosome / antibacterial humoral response / response to lipopolysaccharide / intracellular iron ion homeostasis / early endosome / iron ion binding / response to xenobiotic stimulus / extracellular space / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
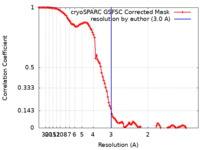
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Newton TP / Zimanyi C / Kopylov M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of Conalbumin from chicken egg white (sigma-Cas 1391-06-6) Authors: Newton TP / Zimanyi C / Kopylov M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29028.map.gz emd_29028.map.gz | 61.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29028-v30.xml emd-29028-v30.xml emd-29028.xml emd-29028.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29028_fsc.xml emd_29028_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_29028.png emd_29028.png | 74.3 KB | ||
| Masks |  emd_29028_msk_1.map emd_29028_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29028.cif.gz emd-29028.cif.gz | 5.7 KB | ||
| Others |  emd_29028_half_map_1.map.gz emd_29028_half_map_1.map.gz emd_29028_half_map_2.map.gz emd_29028_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29028 http://ftp.pdbj.org/pub/emdb/structures/EMD-29028 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29028 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29028 | HTTPS FTP |
-Related structure data
| Related structure data |  8feiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29028.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29028.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.674 Å | ||||||||||||||||||||||||||||||||||||
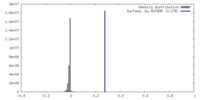
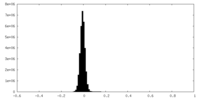
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29028_msk_1.map emd_29028_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
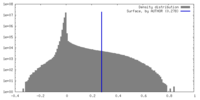
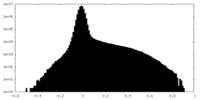
| Density Histograms |
-Half map: Final half map A
| File | emd_29028_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
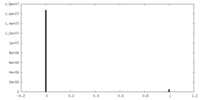
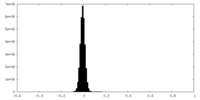
| Density Histograms |
-Half map: Final half map B
| File | emd_29028_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
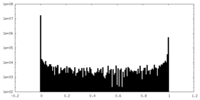
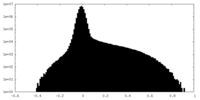
| Density Histograms |
- Sample components
Sample components
-Entire : Conalbumin from chicken egg white
| Entire | Name: Conalbumin from chicken egg white |
|---|---|
| Components |
|
-Supramolecule #1: Conalbumin from chicken egg white
| Supramolecule | Name: Conalbumin from chicken egg white / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Ovotransferrin
| Macromolecule | Name: Ovotransferrin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 77.878531 KDa |
| Sequence | String: MKLILCTVLS LGIAAVCFAA PPKSVIRWCT ISSPEEKKCN NLRDLTQQER ISLTCVQKAT YLDCIKAIAN NEADAISLDG GQAFEAGLA PYKLKPIAAE VYEHTEGSTT SYYAVAVVKK GTEFTVNDLQ GKTSCHTGLG RSAGWNIPIG TLLHRGAIEW E GIESGSVE ...String: MKLILCTVLS LGIAAVCFAA PPKSVIRWCT ISSPEEKKCN NLRDLTQQER ISLTCVQKAT YLDCIKAIAN NEADAISLDG GQAFEAGLA PYKLKPIAAE VYEHTEGSTT SYYAVAVVKK GTEFTVNDLQ GKTSCHTGLG RSAGWNIPIG TLLHRGAIEW E GIESGSVE QAVAKFFSAS CVPGATIEQK LCRQCKGDPK TKCARNAPYS GYSGAFHCLK DGKGDVAFVK HTTVNENAPD QK DEYELLC LDGSRQPVDN YKTCNWARVA AHAVVARDDN KVEDIWSFLS KAQSDFGVDT KSDFHLFGPP GKKDPVLKDL LFK DSAIML KRVPSLMDSQ LYLGFEYYSA IQSMRKDQLT PSPRENRIQW CAVGKDEKSK CDRWSVVSNG DVECTVVDET KDCI IKIMK GEADAVALDG GLVYTAGVCG LVPVMAERYD DESQCSKTDE RPASYFAVAV ARKDSNVNWN NLKGKKSCHT AVGRT AGWV IPMGLIHNRT GTCNFDEYFS EGCAPGSPPN SRLCQLCQGS GGIPPEKCVA SSHEKYFGYT GALRCLVEKG DVAFIQ HST VEENTGGKNK ADWAKNLQMD DFELLCTDGR RANVMDYREC NLAEVPTHAV VVRPEKANKI RDLLERQEKR FGVNGSE KS KFMMFESQNK DLLFKDLTKC LFKVREGTTY KEFLGDKFYT VISSLKTCNP SDILQMCSFL EGK UniProtKB: Ovotransferrin |
-Macromolecule #2: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: PBS |
|---|---|
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 58.53 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)