+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the vertebrate augmin complex | ||||||||||||
 Map data Map data | Sharpened map of X laevis augmin | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | microtubule / branching microtubule nucleation / spindle assembly / CELL CYCLE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationHAUS complex / microtubule minus-end binding / microtubule organizing center organization / mitotic spindle microtubule / microtubule nucleation / centrosome cycle / microtubule organizing center / spindle assembly / bioluminescence / generation of precursor metabolites and energy ...HAUS complex / microtubule minus-end binding / microtubule organizing center organization / mitotic spindle microtubule / microtubule nucleation / centrosome cycle / microtubule organizing center / spindle assembly / bioluminescence / generation of precursor metabolites and energy / microtubule cytoskeleton organization / spindle / spindle pole / mitotic spindle / microtubule binding / microtubule / cell division / centrosome / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species | |||||||||||||
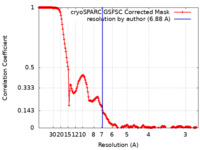
| Method | single particle reconstruction / cryo EM / Resolution: 6.88 Å | ||||||||||||
 Authors Authors | Travis SM / Huang W / Zhang R / Petry S | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Integrated model of the vertebrate augmin complex. Authors: Sophie M Travis / Brian P Mahon / Wei Huang / Meisheng Ma / Michael J Rale / Jodi Kraus / Derek J Taylor / Rui Zhang / Sabine Petry /   Abstract: Accurate segregation of chromosomes is required to maintain genome integrity during cell division. This feat is accomplished by the microtubule-based spindle. To build a spindle rapidly and with high ...Accurate segregation of chromosomes is required to maintain genome integrity during cell division. This feat is accomplished by the microtubule-based spindle. To build a spindle rapidly and with high fidelity, cells take advantage of branching microtubule nucleation, which rapidly amplifies microtubules during cell division. Branching microtubule nucleation relies on the hetero-octameric augmin complex, but lack of structure information about augmin has hindered understanding how it promotes branching. In this work, we combine cryo-electron microscopy, protein structural prediction, and visualization of fused bulky tags via negative stain electron microscopy to identify the location and orientation of each subunit within the augmin structure. Evolutionary analysis shows that augmin's structure is highly conserved across eukaryotes, and that augmin contains a previously unidentified microtubule binding site. Thus, our findings provide insight into the mechanism of branching microtubule nucleation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28981.map.gz emd_28981.map.gz | 266.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28981-v30.xml emd-28981-v30.xml emd-28981.xml emd-28981.xml | 33.1 KB 33.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28981_fsc.xml emd_28981_fsc.xml | 17.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28981.png emd_28981.png | 42.5 KB | ||
| Filedesc metadata |  emd-28981.cif.gz emd-28981.cif.gz | 9.7 KB | ||
| Others |  emd_28981_half_map_1.map.gz emd_28981_half_map_1.map.gz emd_28981_half_map_2.map.gz emd_28981_half_map_2.map.gz | 498.4 MB 498.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28981 http://ftp.pdbj.org/pub/emdb/structures/EMD-28981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28981 | HTTPS FTP |
-Related structure data
| Related structure data |  8fckMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28981.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28981.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of X laevis augmin | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map A of X laevis augmin
| File | emd_28981_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A of X laevis augmin | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B of X laevis augmin
| File | emd_28981_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B of X laevis augmin | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Augmin
| Entire | Name: Augmin |
|---|---|
| Components |
|
-Supramolecule #1: Augmin
| Supramolecule | Name: Augmin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Peak octameric fraction following gel filtration |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 403 KDa |
-Macromolecule #1: HAUS augmin-like complex subunit 1
| Macromolecule | Name: HAUS augmin-like complex subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 32.655621 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDEKSTKIIM WLKKMFGDKP LPPYEVNTRT MEILYQLAEW NEARDKDLSL VTEDLKLKSA EVKAEAKYLQ DLLTEGLGPS YTNLSRMGN NYLNQIVDSC LALELKNSSL SSYIPAVNDL SSELVAIELN NQEMEAELTS LRKKLTEALV LEKSLEQDLK K AEEQCNFE ...String: MDEKSTKIIM WLKKMFGDKP LPPYEVNTRT MEILYQLAEW NEARDKDLSL VTEDLKLKSA EVKAEAKYLQ DLLTEGLGPS YTNLSRMGN NYLNQIVDSC LALELKNSSL SSYIPAVNDL SSELVAIELN NQEMEAELTS LRKKLTEALV LEKSLEQDLK K AEEQCNFE KAKVEIRSQN MKKLKDKSEE YKYKIHAAKD QLSSAGMEEP LTHRSLVSLS ETLTELKAQS MAAKEKLNSY LD LAPNPSL VKVKIEEAKR ELKATEVELT TKVNMMEFVV PEPSKRRLK UniProtKB: HAUS augmin-like complex subunit 1 |
-Macromolecule #2: HAUS augmin-like complex subunit 3
| Macromolecule | Name: HAUS augmin-like complex subunit 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 68.11225 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGGDRFVQT LQKLNYPKGA QLDGEDFDWL FEAVDLKPFL DWFCSAASEQ NVVPDEKLQA FNTLKESGKP VLDEKALDEV LKTFSISKV PAIEEVAIEK LEEEVKALQK QKNLHIRRRN KLQMVESGNR QMCLKSKDKE EETGRAFQEV LHLLRVTNKK L NHELQSIV ...String: MSGGDRFVQT LQKLNYPKGA QLDGEDFDWL FEAVDLKPFL DWFCSAASEQ NVVPDEKLQA FNTLKESGKP VLDEKALDEV LKTFSISKV PAIEEVAIEK LEEEVKALQK QKNLHIRRRN KLQMVESGNR QMCLKSKDKE EETGRAFQEV LHLLRVTNKK L NHELQSIV NGVQTLMSFF STPETACELS SQPIFLSQLL LDKYLSLEEQ STAALTSFTK EHFFEGMSKF VEGSDENFQL VQ LNVNSFG EDGTTEDKCK EMMRLQLAYI CAKHKLIQMK AKSASLKVGL QWAENNASVV QDKASQKEEN LKVRITSLKN ETL QIENHT NSISNEKLPG LVRDNAQLLN MPIVKGDYDL QMAHQTSCSS RQDLVCDHLM KQKASFELLQ LGYELELRKH RDVY RELGS IVQELKESGD KLEERLTMLS DVNLLSASKP RSNIDSKDLT SHRLYQLLDG DNTQKLFRTY DGLESVAQKL SQDIA SMRD QLEVSEQEHS LLLSKLDSHL KELRDFMYPE GNTLMLTTPE LSGEFHQLGS QLEKLNHITV EILGDLQLKR KMLESN KLQ QIEKQLYVYF FQNEEQLKSI VGKLEAQTGG GSSA UniProtKB: HAUS augmin-like complex subunit 3 |
-Macromolecule #3: HAUS augmin like complex subunit 4 L homeolog
| Macromolecule | Name: HAUS augmin like complex subunit 4 L homeolog / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 41.256438 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAQTLQYVSS RLSMLQIDEE DLERNAQFGK VLIELCPLLG PNGGSANLNR ELEETRRELL LQRKMWMRSE VIYQLVQEML LDLQVRKLE GSLTEEERKF QDGLQQCMLV SECSRLLTAD SVPPSDSTSI LGLDKQDLLD LLPPNMLVLW VRDRLQKQLE E ALKKKCFT ...String: MAQTLQYVSS RLSMLQIDEE DLERNAQFGK VLIELCPLLG PNGGSANLNR ELEETRRELL LQRKMWMRSE VIYQLVQEML LDLQVRKLE GSLTEEERKF QDGLQQCMLV SECSRLLTAD SVPPSDSTSI LGLDKQDLLD LLPPNMLVLW VRDRLQKQLE E ALKKKCFT FLSFHQPETD EEGDVLRAAK VLRLASTLED EKRRLQNEQE KHQEMRALLE KQQEIYPHVL LRCLSLLRQA AS ELRLRAQ SDIDRINAEY LEAKSNALFL KLRMEELQVL TDCYTPEKVL VHRQIRDTLE AGVKKEKQEL STSRQILSSY EFL GPEFEG LVQEYTRLKD KIKDNRWMLQ ELSKSLP UniProtKB: HAUS augmin like complex subunit 4 L homeolog |
-Macromolecule #4: HAUS augmin-like complex subunit 5
| Macromolecule | Name: HAUS augmin-like complex subunit 5 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 77.357281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MERRSLAQEL KKWAVEEMGL PAQKAPSEEM LQRLFIGQCG DIWKFIIRHI HSHRTVRKIE GNLLWYQQLQ HTEAQRTAEE EQQQRRKQL CKEILELRAE LHHLQEQIQT AEREIVGQDL NCERAQDLCR RSLLLRAFNK KREEECEALC QSNKKIQYRC E QLQEIRRA ...String: MERRSLAQEL KKWAVEEMGL PAQKAPSEEM LQRLFIGQCG DIWKFIIRHI HSHRTVRKIE GNLLWYQQLQ HTEAQRTAEE EQQQRRKQL CKEILELRAE LHHLQEQIQT AEREIVGQDL NCERAQDLCR RSLLLRAFNK KREEECEALC QSNKKIQYRC E QLQEIRRA SQREVMFSAV DPDLSSSTFL EPEVLRDVRE VCKLRFKFLR SLHDDSISSS VHPGKEDLRS LSHQQWMSMA EK VWNTHTP NHILAALERL TLNSTQELKK LQFSQAADLS KGPSCQLKEF SEPITQSRSC NESTHLDPQE TLPSFHSLIQ EGW ANSVKV SSELRRVQSQ AQALSEHLAE RIQEIHKKLS DGSEVSVLTR AAFDAELRCV ILRGCRDALM QECRMLQEEA AGKK QEMKL LQQQQQNIQE ACLLLDKKQK HIQILIKGNS SSKSQIRRSS VEAQKYVQDK LLPWPQEIIQ ESQRLQDSIQ KEVKH FSAI CLPALLKVST DGFNLLPSRE LSINRMSNTH APYYGIFKGI YESVRLPLYK APESVLSHVA DMKKQLFFLR SQLSSR SEA ISKTQRALQK NTNPDTDALL KSLSDHYSLE LDEMVPKMQR LIQQCEKHQE YGKEVQATVM DWWEQPVQLC LPSEERG GL TLRQWRERWT VAVTALQRAT GSRS UniProtKB: HAUS augmin-like complex subunit 5 |
-Macromolecule #5: HAUS augmin like complex subunit 2 L homeolog, Green fluorescent ...
| Macromolecule | Name: HAUS augmin like complex subunit 2 L homeolog, Green fluorescent protein chimera type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 53.505473 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMAANPWSPV QPSAASLLLE KCLAAGVLSQ NALDQAQLEA PCFSHAEELQ RITEIKAEIN QKSLELELLR LEKETADITH PLYLGQKHQ ALQAMNSHLE AVLREKRSLR QRLAQPVCQE NLPIEASYHR FTAELLPLAV NFIEKLEIYI KTIQTIPKIP D CASNMDNA ...String: MMAANPWSPV QPSAASLLLE KCLAAGVLSQ NALDQAQLEA PCFSHAEELQ RITEIKAEIN QKSLELELLR LEKETADITH PLYLGQKHQ ALQAMNSHLE AVLREKRSLR QRLAQPVCQE NLPIEASYHR FTAELLPLAV NFIEKLEIYI KTIQTIPKIP D CASNMDNA LMRMESLEAE MEEVTEQILT WREQQKTAFQ MNSDANSSCI TAQTSYLSIE NLHPVDMVSK GEELFTGVVP IL VELDGDV NGHKFSVSGE GEGDATYGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKQHD FFKSAMPEGY VQE RTIFFK DDGNYKTRAE VKFEGDTLVN RIELKGIDFK EDGNILGHKL EYNYNSHNVY IMADKQKNGI KVNFKIRHNI EDGS VQLAD HYQQNTPIGD GPVLLPDNHY LSTQSALSKD PNEKRDHMVL KEFVTAAGIT LGMDELYKVD HHHHHHH UniProtKB: HAUS augmin like complex subunit 2 L homeolog, Green fluorescent protein |
-Macromolecule #6: HAUS augmin like complex subunit 6 L homeolog
| Macromolecule | Name: HAUS augmin like complex subunit 6 L homeolog / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 50.927965 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPASGSTRGA EFMQSGSRPH LAWQREHMWL ALQGLGFESG AEAANAGKTL VHVTFGVNMF DKPNKDAFYV VFHFLFGKLD NVRCKEVFR YCWPPLDKKR DAEFRKACCE WLKKISDEVG AGFPQVVASI FLSPGGPKFV HLLYHFARYV MLQHIKRDAD A GNVFISEA ...String: GPASGSTRGA EFMQSGSRPH LAWQREHMWL ALQGLGFESG AEAANAGKTL VHVTFGVNMF DKPNKDAFYV VFHFLFGKLD NVRCKEVFR YCWPPLDKKR DAEFRKACCE WLKKISDEVG AGFPQVVASI FLSPGGPKFV HLLYHFARYV MLQHIKRDAD A GNVFISEA LQSKIQDPQK ALARNKLARQ KYLKVLQKEN LVIEEYQRKA QLLIKQIRDM RSEHVALQNQ QKLAEKVDRK IS DKDENIQ KTRCMWNTIM QMLKEMEKEV DVVDAVVRGN IDQYCLDGTN ATLNIPNLLI SRIESEMHRL QMDNVYEAGK VNL ITVVQL LNEALKLVSG ERSLYDCKGV RLDLQYLHGK AKFESEVLTR LRNMRHKIKR EDLVSIEKII ADREREWERK WEKI LGKCP FSLLKGLNPA LELNPPMAPF SFDPASEEVL KSSVFCH UniProtKB: HAUS augmin like complex subunit 6 L homeolog |
-Macromolecule #7: HAUS augmin like complex subunit 7 S homeolog
| Macromolecule | Name: HAUS augmin like complex subunit 7 S homeolog / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 39.379605 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTGGKELGAA VELYERLQML SCPCLEGVYL TDPQSIYELL CTPSSHRLDI LQWLCSRIYP PVQEQLSSLK ESQTDTKVKE IAKLCFDLM LCHFDDLDLI RGHASPFKQI SFIGQLLDVI QYPDTISSNV ILESLSHSTE KNVVTCIREN EELLKELFSS P HFQATLSP ...String: MTGGKELGAA VELYERLQML SCPCLEGVYL TDPQSIYELL CTPSSHRLDI LQWLCSRIYP PVQEQLSSLK ESQTDTKVKE IAKLCFDLM LCHFDDLDLI RGHASPFKQI SFIGQLLDVI QYPDTISSNV ILESLSHSTE KNVVTCIREN EELLKELFSS P HFQATLSP ECNPWPADFK PLLNAEESLQ KRATQSSKGK DMSNSVEALL EISSSLKALK EECVDLCSSV TDGDKVIQSL RL ALTDFHQ LTIAFNQIYA NEFQEHCGHP APHMSPMGPF FQFVHQSLST CFKELESIAQ FTETSENIVD VVRERHQSKE KWA GSTIST LCEKMKELRQ SYEAFQQSSL QD UniProtKB: HAUS augmin like complex subunit 7 S homeolog |
-Macromolecule #8: HAUS augmin-like complex subunit 8
| Macromolecule | Name: HAUS augmin-like complex subunit 8 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 41.144852 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSEAGVAPIE DGSQNSSGGS SGDAALKKSK GGAKVVKSRY MQIGRSKVSK NSLANTTVCS GGKVPERGSG GTPTRRSLAP HKAKITAAV PLPALDGSIF TKEDLQSTLL DGHRIARPDL DLSVINDRTL QKITPRPVVT SEQKKPKRDT TPVNLVPEDM V EMIESQTL ...String: MSEAGVAPIE DGSQNSSGGS SGDAALKKSK GGAKVVKSRY MQIGRSKVSK NSLANTTVCS GGKVPERGSG GTPTRRSLAP HKAKITAAV PLPALDGSIF TKEDLQSTLL DGHRIARPDL DLSVINDRTL QKITPRPVVT SEQKKPKRDT TPVNLVPEDM V EMIESQTL LLTYLTIKMQ KNLFRLEEKA ERNLLLVNDQ KDQLQETIHM MKRDLTLLQR EERLRDLIEK QDEVLTPVVT SK DPFKDNY TTFATALDST RHQLAIKNIH ITGNRHRYLE ELQKHLAITK SLLEEIMPSH ASENAESFDT IKDLENIVLK TDE ELARSF RQILDLSFKV NKEISLQSQK AVEETCESAL VRQWYFDGSL P UniProtKB: HAUS augmin-like complex subunit 8 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.06 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.7 Component:
| ||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: HOLEY / Support film - #0 - Film thickness: 10 / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: CONTINUOUS / Support film - #1 - Film thickness: 5 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 8 sec. | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 2350 / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 39.903 µm / Nominal defocus min: 10.105 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-8fck: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)