[English] 日本語
 Yorodumi
Yorodumi- EMDB-28959: DNA replication fork binding triggers structural changes in the P... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | |||||||||
 Map data Map data | final sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PriA / PriB / replication restart / E. coli / HELICASE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpre-primosome complex / DnaB-DnaC-DnaT-PriA-PriC complex / DnaB-DnaC-DnaT-PriA-PriB complex / primosome complex / plasmid maintenance / DNA replication, synthesis of primer / 3'-5' DNA helicase activity / DNA 3'-5' helicase / replication fork processing / DNA replication initiation ...pre-primosome complex / DnaB-DnaC-DnaT-PriA-PriC complex / DnaB-DnaC-DnaT-PriA-PriB complex / primosome complex / plasmid maintenance / DNA replication, synthesis of primer / 3'-5' DNA helicase activity / DNA 3'-5' helicase / replication fork processing / DNA replication initiation / response to gamma radiation / response to radiation / helicase activity / DNA-templated DNA replication / double-strand break repair / single-stranded DNA binding / DNA recombination / DNA replication / response to antibiotic / ATP hydrolysis activity / DNA binding / zinc ion binding / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
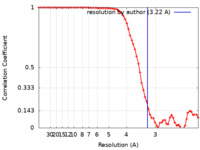
| Method | single particle reconstruction / cryo EM / Resolution: 3.22 Å | |||||||||
 Authors Authors | Duckworth AT / Ducos PL / McMillan SD / Satyshur KA / Blumenthal KH / Deorio HR / Larson JA / Sandler SJ / Grant T / Keck JL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli. Authors: Alexander T Duckworth / Peter L Ducos / Sarah D McMillan / Kenneth A Satyshur / Katelien H Blumenthal / Haley R Deorio / Joseph A Larson / Steven J Sandler / Timothy Grant / James L Keck /  Abstract: Bacterial replisomes often dissociate from replication forks before chromosomal replication is complete. To avoid the lethal consequences of such situations, bacteria have evolved replication restart ...Bacterial replisomes often dissociate from replication forks before chromosomal replication is complete. To avoid the lethal consequences of such situations, bacteria have evolved replication restart pathways that reload replisomes onto prematurely terminated replication forks. To understand how the primary replication restart pathway in E. coli (PriA-PriB) selectively acts on replication forks, we determined the cryogenic-electron microscopy structure of a PriA/PriB/replication fork complex. Replication fork specificity arises from extensive PriA interactions with each arm of the branched DNA. These interactions reshape the PriA protein to create a pore encircling single-stranded lagging-strand DNA while also exposing a surface of PriA onto which PriB docks. Together with supporting biochemical and genetic studies, the structure reveals a switch-like mechanism for replication restart initiation in which restructuring of PriA directly couples replication fork recognition to PriA/PriB complex formation to ensure robust and high-fidelity replication re-initiation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28959.map.gz emd_28959.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28959-v30.xml emd-28959-v30.xml emd-28959.xml emd-28959.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28959_fsc.xml emd_28959_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_28959.png emd_28959.png | 122.1 KB | ||
| Filedesc metadata |  emd-28959.cif.gz emd-28959.cif.gz | 6.9 KB | ||
| Others |  emd_28959_half_map_1.map.gz emd_28959_half_map_1.map.gz emd_28959_half_map_2.map.gz emd_28959_half_map_2.map.gz | 25 MB 25 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28959 http://ftp.pdbj.org/pub/emdb/structures/EMD-28959 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28959 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28959 | HTTPS FTP |
-Related structure data
| Related structure data |  8fakMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28959.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28959.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.079 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: unfiltered / unsharpened half map 1
| File | emd_28959_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered / unsharpened half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: unfiltered / unsharpened half map 2
| File | emd_28959_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered / unsharpened half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of PriA, PriB, and replication fork
| Entire | Name: Complex of PriA, PriB, and replication fork |
|---|---|
| Components |
|
-Supramolecule #1: Complex of PriA, PriB, and replication fork
| Supramolecule | Name: Complex of PriA, PriB, and replication fork / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134 KDa |
-Macromolecule #1: Primosomal replication protein N
| Macromolecule | Name: Primosomal replication protein N / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.459194 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTNRLVLSGT VCRAPLRKVS PSGIPHCQFV LEHRSVQEEA GFHRQAWCQM PVIVSGHENQ AITHSITVGS RITVQGFISC HKAKNGLSK MVLHAEQIEL IDSGD UniProtKB: Replication restart protein PriB |
-Macromolecule #3: Primosomal protein N'
| Macromolecule | Name: Primosomal protein N' / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81.76582 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPVAHVALPV PLPRTFDYLL PEGMTVKAGC RVRVPFGKQQ ERIGIVVSVS DASELPLNEL KAVVEVLDSE PVFTHSVWRL LLWAADYYH HPIGDVLFHA LPILLRQGRP AANAPMWYWF ATEQGQAVDL NSLKRSPKQQ QALAALRQGK IWRDQVATLE F NDAALQAL ...String: MPVAHVALPV PLPRTFDYLL PEGMTVKAGC RVRVPFGKQQ ERIGIVVSVS DASELPLNEL KAVVEVLDSE PVFTHSVWRL LLWAADYYH HPIGDVLFHA LPILLRQGRP AANAPMWYWF ATEQGQAVDL NSLKRSPKQQ QALAALRQGK IWRDQVATLE F NDAALQAL RKKGLCDLAS ETPEFSDWRT NYAVSGERLR LNTEQATAVG AIHSAADTFS AWLLAGVTGS GKTEVYLSVL EN VLAQGKQ ALVMVPEIGL TPQTIARFRE RFNAPVEVLH SGLNDSERLS AWLKAKNGEA AIVIGTRSAL FTPFKNLGVI VID EEHDSS YKQQEGWRYH ARDLAVYRAH SEQIPIILGS ATPALETLCN VQQKKYRLLR LTRRAGNARP AIQHVLDLKG QKVQ AGLAP ALITRMRQHL QADNQVILFL NRRGFAPALL CHDCGWIAEC PRCDHYYTLH QAQHHLRCHH CDSQRPVPRQ CPSCG STHL VPVGLGTEQL EQTLAPLFPG VPISRIDRDT TSRKGALEQQ LAEVHRGGAR ILIGTQMLAK GHHFPDVTLV ALLDVD GAL FSADFRSAER FAQLYTQVAG RAGRAGKQGE VVLQTHHPEH PLLQTLLYKG YDAFAEQALA ERRMMQLPPW TSHVIVR AE DHNNQHAPLF LQQLRNLILS SPLADEKLWV LGPVPALAPK RGGRWRWQIL LQHPSRVRLQ HIINGTLALI NTIPDSRK V KWVLDVDPIE G UniProtKB: Replication restart protein PriA |
-Macromolecule #2: DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP...
| Macromolecule | Name: DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3') type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 12.206812 KDa |
| Sequence | String: (DG)(DC)(DC)(DG)(DC)(DA)(DG)(DA)(DC)(DT) (DC)(DA)(DT)(DT)(DT)(DA)(DG)(DC)(DC)(DC) (DT)(DT)(DA)(DT)(DC)(DC)(DG)(DT)(DA) (DT)(DT)(DG)(DC)(DG)(DG)(DT)(DC)(DT)(DC) (DG) |
-Macromolecule #4: DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP...
| Macromolecule | Name: DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 12.399983 KDa |
| Sequence | String: (DC)(DG)(DA)(DG)(DA)(DC)(DC)(DG)(DC)(DA) (DA)(DT)(DA)(DC)(DG)(DG)(DA)(DT)(DA)(DA) (DG)(DG)(DG)(DC)(DT)(DG)(DA)(DG)(DC) (DA)(DC)(DG)(DC)(DC)(DG)(DA)(DC)(DG)(DA) (DA) |
-Macromolecule #5: DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3') / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.567944 KDa |
| Sequence | String: (DT)(DT)(DC)(DG)(DT)(DC)(DG)(DG)(DC)(DG) (DT)(DG)(DC)(DT)(DC) |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Number grids imaged: 1 / #0 - Number real images: 1600 / #0 - Average electron dose: 100.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: FEI FALCON III (4k x 4k) / #1 - Detector mode: INTEGRATING / #1 - Number grids imaged: 1 / #1 - Number real images: 1200 / #1 - Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8fak: |
 Movie
Movie Controller
Controller



 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)