+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the LRRK1 dimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | kinase / LRRK / multi-domain protein / GTPase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationosteoclast development / positive regulation of intracellular signal transduction / bone resorption / positive regulation of canonical Wnt signaling pathway / non-specific serine/threonine protein kinase / intracellular signal transduction / protein serine kinase activity / protein serine/threonine kinase activity / GTP binding / mitochondrion ...osteoclast development / positive regulation of intracellular signal transduction / bone resorption / positive regulation of canonical Wnt signaling pathway / non-specific serine/threonine protein kinase / intracellular signal transduction / protein serine kinase activity / protein serine/threonine kinase activity / GTP binding / mitochondrion / ATP binding / metal ion binding / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
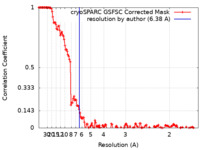
| Method | single particle reconstruction / cryo EM / Resolution: 6.38 Å | |||||||||
 Authors Authors | Metcalfe RD / Martinez Fiesco JA / Zhang P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure and regulation of full-length human leucine-rich repeat kinase 1. Authors: Riley D Metcalfe / Juliana A Martinez Fiesco / Luis Bonet-Ponce / Jillian H Kluss / Mark R Cookson / Ping Zhang /  Abstract: The human leucine-rich repeat kinases (LRRKs), LRRK1 and LRRK2 are large and unusually complex multi-domain kinases, which regulate fundamental cellular processes and have been implicated in human ...The human leucine-rich repeat kinases (LRRKs), LRRK1 and LRRK2 are large and unusually complex multi-domain kinases, which regulate fundamental cellular processes and have been implicated in human disease. Structures of LRRK2 have recently been determined, but the structure and molecular mechanisms regulating the activity of the LRRK1 as well as differences in the regulation of LRRK1 and LRRK2 remain unclear. Here, we report a cryo-EM structure of the LRRK1 monomer and a lower-resolution cryo-EM map of the LRRK1 dimer. The monomer structure, in which the kinase is in an inactive conformation, reveals key interdomain interfaces that control kinase activity as we validate experimentally. Both the LRRK1 monomer and dimer are structurally distinct compared to LRRK2. Overall, our results provide structural insights into the activation of the human LRRKs, which advance our understanding of their physiological and pathological roles. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28952.map.gz emd_28952.map.gz | 288.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28952-v30.xml emd-28952-v30.xml emd-28952.xml emd-28952.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28952_fsc.xml emd_28952_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28952.png emd_28952.png | 71 KB | ||
| Masks |  emd_28952_msk_1.map emd_28952_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Others |  emd_28952_additional_1.map.gz emd_28952_additional_1.map.gz emd_28952_half_map_1.map.gz emd_28952_half_map_1.map.gz emd_28952_half_map_2.map.gz emd_28952_half_map_2.map.gz | 146.2 MB 285 MB 285 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28952 http://ftp.pdbj.org/pub/emdb/structures/EMD-28952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28952 | HTTPS FTP |
-Related structure data
| Related structure data |  8facC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28952.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28952.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28952_msk_1.map emd_28952_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_28952_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28952_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_28952_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Leucine-rich repeat kinase 1
| Entire | Name: Leucine-rich repeat kinase 1 |
|---|---|
| Components |
|
-Supramolecule #1: Leucine-rich repeat kinase 1
| Supramolecule | Name: Leucine-rich repeat kinase 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: Leucine-rich repeat kinase 1
| Macromolecule | Name: Leucine-rich repeat kinase 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDHDGDY KDHDIDYKDD DDKLGLEVLF QGPMAGMSQR PPSMYWCVGP EESAVCPERA METLNGAGDT GGKPSTRGGD PAARSRRTEG IRAAYRRGDR GGARDLLEEA CDQCASQLEK GQLLSIPAAY GDLEMVRYLL SKRLVELPTE PTDDNPAVVA AYFGHTAVVQ ...String: MDYKDHDGDY KDHDIDYKDD DDKLGLEVLF QGPMAGMSQR PPSMYWCVGP EESAVCPERA METLNGAGDT GGKPSTRGGD PAARSRRTEG IRAAYRRGDR GGARDLLEEA CDQCASQLEK GQLLSIPAAY GDLEMVRYLL SKRLVELPTE PTDDNPAVVA AYFGHTAVVQ ELLESLPGPC SPQRLLNWML ALACQRGHLG VVKLLVLTHG ADPESYAVRK NEFPVIVRLP LYAAIKSGNE DIAIFLLRHG AYFCSYILLD SPDPSKHLLR KYFIEASPLP SSYPGKTALR VKWSHLRLPW VDLDWLIDIS CQITELDLSA NCLATLPSVI PWGLINLRKL NLSDNHLGEL PGVQSSDEII CSRLLEIDIS SNKLSHLPPG FLHLSKLQKL TASKNCLEKL FEEENATNWI GLRKLQELDI SDNKLTELPA LFLHSFKSLN SLNVSRNNLK VFPDPWACPL KCCKASRNAL ECLPDKMAVF WKNHLKDVDF SENALKEVPL GLFQLDALMF LRLQGNQLAA LPPQEKWTCR QLKTLDLSRN QLGKNEDGLK TKRIAFFTTR GRQRSGTEAA SVLEFPAFLS ESLEVLCLND NHLDTVPPSV CLLKSLSELY LGNNPGLREL PPELGQLGNL WQLDTEDLTI SNVPAEIQKE GPKAMLSYLR AQLRKAEKCK LMKMIIVGPP RQGKSTLLEI LQTGRAPQVV HGEATIRTTK WELQRPAGSR AKVESVEFNV WDIGGPASMA TVNQCFFTDK ALYVVVWNLA LGEEAVANLQ FWLLNIEAKA PNAVVLVVGT HLDLIEAKFR VERIATLRAY VLALCRSPSG SRATGFPDIT FKHLHEISCK SLEGQEGLRQ LIFHVTCSMK DVGSTIGCQR LAGRLIPRSY LSLQEAVLAE QQRRSRDDDV QYLTDRQLEQ LVEQTPDNDI KDYEDLQSAI SFLIETGTLL HFPDTSHGLR NLYFLDPIWL SECLQRIFNI KGSRSVAKNG VIRAEDLRML LVGTGFTQQT EEQYFQFLAK FEIALPVAND SYLLPHLLPS KPGLDTHGMR HPTANTIQRV FKMSFVPVGF WQRFIARMLI SLAEMDLQLF ENKKNTKSRN RKVTIYSFTG NQRNRCSTFR VKRNQTIYWQ EGLLVTFDGG YLSVESSDVN WKKKKSGGMK IVCQSEVRDF SAMAFITDHV NSLIDQWFPA LTATESDGTP LMEQYVPCPV CETAWAQHTD PSEKSEDVQY FDMEDCVLTA IERDFISCPR HPDLPVPLQE LVPELFMTDF PARLFLENSK LEHSEDEGSV LGQGGSGTVI YRARYQGQPV AVKRFHIKKF KNFANVPADT MLRHLRATDA MKNFSEFRQE ASMLHALQHP CIVALIGISI HPLCFALELA PLSSLNTVLS ENARDSSFIP LGHMLTQKIA YQIASGLAYL HKKNIIFCDL KSDNILVWSL DVKEHINIKL SDYGISRQSF HEGALGVEGT PGYQAPEIRP RIVYDEKVDM FSYGMVLYEL LSGQRPALGH HQLQIAKKLS KGIRPVLGQP EEVQFRRLQA LMMECWDTKP EKRPLALSVV SQMKDPTFAT FMYELCCGKQ TAFFSSQGQE YTVVFWDGKE ESRNYTVVNT EKGLMEVQRM CCPGMKVSCQ LQVQRSLWTA TEDQKIYIYT LKGMCPLNTP QQALDTPAVV TCFLAVPVIK KNSYLVLAGL ADGLVAVFPV VRGTPKDSCS YLCSHTANRS KFSIADEDAR QNPYPVKAME VVNSGSEVWY SNGPGLLVID CASLEICRRL EPYMAPSMVT SVVCSSEGRG EEVVWCLDDK ANSLVMYHST TYQLCARYFC GVPSPLRDMF PVRPLDTEPP AASHTANPKV PEGDSIADVS IMYSEELGTQ ILIHQESLTD YCSMSSYSSS PPRQAARSPS SLPSSPASSS SVPFSTDCED SDMLHTPGAA SDRSEHDLTP MDGETFSQHL QAVKILAVRD LIWVPRRGGD VIVIGLEKDS GAQRGRVIAV LKARELTPHG VLVDAAVVAK DTVVCTFENE NTEWCLAVWR GWGAREFDIF YQSYEELGRL EACTRKRR UniProtKB: Leucine-rich repeat serine/threonine-protein kinase 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 8.3 |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
| Details | Purified LRRK1 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 7 / Number real images: 18074 / Average exposure time: 2.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)