登録情報 データベース : EMDB / ID : EMD-28915タイトル SIRT6 bound to an H3K9Ac nucleosome map A 複合体 : SIRT6 bound to H3K9Acタンパク質・ペプチド : Histone H3.2タンパク質・ペプチド : Histone H4タンパク質・ペプチド : Histone H2A type 1タンパク質・ペプチド : Histone H2BDNA : DNA (148-MER)DNA : DNA (148-MER)タンパク質・ペプチド : NAD-dependent protein deacylase sirtuin-6リガンド : [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(3aR,5R,6R,6aR)-6-hydroxytetrahydro-2H-furo[2,3-d][1,3]oxathiol-5-yl]methyl dihydrogen diphosphate (non-preferred name)リガンド : ZINC ION / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
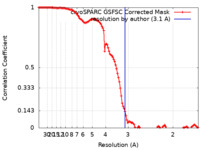
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Xenopus laevis (アフリカツメガエル) / Homo sapiens (ヒト) / synthetic construct (人工物) 手法 / / 解像度 : 3.1 Å Markert J / Whedon S / Wang Z / Cole P / Farnung L 資金援助 1件 Organization Grant number 国 Not funded
ジャーナル : J Am Chem Soc / 年 : 2023タイトル : Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.著者 : Zhipeng A Wang / Jonathan W Markert / Samuel D Whedon / Maheeshi Yapa Abeywardana / Kwangwoon Lee / Hanjie Jiang / Carolay Suarez / Hening Lin / Lucas Farnung / Philip A Cole / 要旨 : The reversible acetylation of histone lysine residues is controlled by the action of acetyltransferases and deacetylases (HDACs), which regulate chromatin structure and gene expression. The sirtuins ... The reversible acetylation of histone lysine residues is controlled by the action of acetyltransferases and deacetylases (HDACs), which regulate chromatin structure and gene expression. The sirtuins are a family of NAD-dependent HDAC enzymes, and one member, sirtuin 6 (Sirt6), influences DNA repair, transcription, and aging. Here, we demonstrate that Sirt6 is efficient at deacetylating several histone H3 acetylation sites, including its canonical site Lys9, in the context of nucleosomes but not free acetylated histone H3 protein substrates. By installing a chemical warhead at the Lys9 position of histone H3, we trap a catalytically poised Sirt6 in complex with a nucleosome and employ this in cryo-EM structural analysis. The structure of Sirt6 bound to a nucleosome reveals extensive interactions between distinct segments of Sirt6 and the H2A/H2B acidic patch and nucleosomal DNA, which accounts for the rapid deacetylation of nucleosomal H3 sites and the disfavoring of histone H2B acetylation sites. These findings provide a new framework for understanding how HDACs target and regulate chromatin. 履歴 登録 2022年11月21日 - ヘッダ(付随情報) 公開 2023年4月5日 - マップ公開 2023年4月5日 - 更新 2025年5月21日 - 現状 2025年5月21日 処理サイト : RCSB / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報
 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト) / synthetic construct (人工物)
Homo sapiens (ヒト) / synthetic construct (人工物) データ登録者
データ登録者 引用
引用 ジャーナル: J Am Chem Soc / 年: 2023
ジャーナル: J Am Chem Soc / 年: 2023
 構造の表示
構造の表示 ダウンロードとリンク
ダウンロードとリンク emd_28915.map.gz
emd_28915.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-28915-v30.xml
emd-28915-v30.xml emd-28915.xml
emd-28915.xml EMDBヘッダ
EMDBヘッダ emd_28915_fsc.xml
emd_28915_fsc.xml FSCデータファイル
FSCデータファイル emd_28915.png
emd_28915.png emd_28915_msk_1.map
emd_28915_msk_1.map マスクマップ
マスクマップ emd-28915.cif.gz
emd-28915.cif.gz emd_28915_additional_1.map.gz
emd_28915_additional_1.map.gz emd_28915_additional_2.map.gz
emd_28915_additional_2.map.gz emd_28915_additional_3.map.gz
emd_28915_additional_3.map.gz emd_28915_additional_4.map.gz
emd_28915_additional_4.map.gz emd_28915_half_map_1.map.gz
emd_28915_half_map_1.map.gz emd_28915_half_map_2.map.gz
emd_28915_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-28915
http://ftp.pdbj.org/pub/emdb/structures/EMD-28915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28915
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28915 emd_28915_validation.pdf.gz
emd_28915_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_28915_full_validation.pdf.gz
emd_28915_full_validation.pdf.gz emd_28915_validation.xml.gz
emd_28915_validation.xml.gz emd_28915_validation.cif.gz
emd_28915_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28915
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28915 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28915
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28915
 F&H 検索
F&H 検索 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_28915.map.gz / 形式: CCP4 / 大きさ: 163.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_28915.map.gz / 形式: CCP4 / 大きさ: 163.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_28915_msk_1.map
emd_28915_msk_1.map 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析
 ムービー
ムービー コントローラー
コントローラー








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)