[English] 日本語
 Yorodumi
Yorodumi- EMDB-28870: CryoEM structure of Arabidopsis thaliana phytochrome A (consensus map) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Arabidopsis thaliana phytochrome A (consensus map) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Phytochrome / asymmetric dimer / gene regulation | |||||||||
| Biological species |  | |||||||||
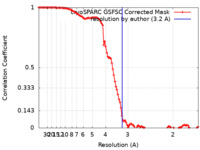
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2023 Journal: Nat Plants / Year: 2023Title: The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms. Authors: E Sethe Burgie / Hua Li / Zira T K Gannam / Katrice E McLoughlin / Richard D Vierstra / Huilin Li /  Abstract: Plants employ a divergent cohort of phytochrome (Phy) photoreceptors to govern many aspects of morphogenesis through reversible photointerconversion between inactive Pr and active Pfr conformers. The ...Plants employ a divergent cohort of phytochrome (Phy) photoreceptors to govern many aspects of morphogenesis through reversible photointerconversion between inactive Pr and active Pfr conformers. The two most influential are PhyA whose retention of Pfr enables sensation of dim light, while the relative instability of Pfr for PhyB makes it better suited for detecting full sun and temperature. To better understand these contrasts, we solved, by cryo-electron microscopy, the three-dimensional structure of full-length PhyA as Pr. Like PhyB, PhyA dimerizes through head-to-head assembly of its C-terminal histidine kinase-related domains (HKRDs), while the remainder assembles as a head-to-tail light-responsive platform. Whereas the platform and HKRDs associate asymmetrically in PhyB dimers, these lopsided connections are absent in PhyA. Analysis of truncation and site-directed mutants revealed that this decoupling and altered platform assembly have functional consequences for Pfr stability of PhyA and highlights how plant Phy structural diversification has extended light and temperature perception. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28870.map.gz emd_28870.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28870-v30.xml emd-28870-v30.xml emd-28870.xml emd-28870.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28870_fsc.xml emd_28870_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28870.png emd_28870.png | 127.4 KB | ||
| Others |  emd_28870_half_map_1.map.gz emd_28870_half_map_1.map.gz emd_28870_half_map_2.map.gz emd_28870_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28870 http://ftp.pdbj.org/pub/emdb/structures/EMD-28870 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28870 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28870 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28870.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28870.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_28870_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
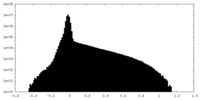
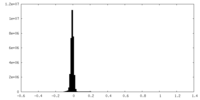
| Density Histograms |
-Half map: #2
| File | emd_28870_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
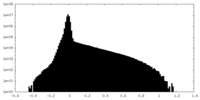
| Density Histograms |
- Sample components
Sample components
-Entire : phytochrome A
| Entire | Name: phytochrome A |
|---|---|
| Components |
|
-Supramolecule #1: phytochrome A
| Supramolecule | Name: phytochrome A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 249 KDa |
-Macromolecule #1: Arabidopsis thaliana phytochrome A
| Macromolecule | Name: Arabidopsis thaliana phytochrome A / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: GMSGSRPTQS SEGSRRSRHS ARIIAQTTVD AKLHADFEES GSSFDYSTSV RVTGPVVENQ PPRSDKVTTT YLHHIQKGKL IQPFGCLLAL DEKTFKVIAY SENASELLTM ASHAVPSVGE HPVLGIGTDI RSLFTAPSAS ALQKALGFGD VSLLNPILVH CRTSAKPFYA ...String: GMSGSRPTQS SEGSRRSRHS ARIIAQTTVD AKLHADFEES GSSFDYSTSV RVTGPVVENQ PPRSDKVTTT YLHHIQKGKL IQPFGCLLAL DEKTFKVIAY SENASELLTM ASHAVPSVGE HPVLGIGTDI RSLFTAPSAS ALQKALGFGD VSLLNPILVH CRTSAKPFYA IIHRVTGSII IDFEPVKPYE VPMTAAGALQ SYKLAAKAIT RLQSLPSGSM ERLCDTMVQE VFELTGYDRV MAYKFHEDDH GEVVSEVTKP GLEPYLGLHY PATDIPQAAR FLFMKNKVRM IVDCNAKHAR VLQDEKLSFD LTLCGSTLRA PHSCHLQYMA NMDSIASLVM AVVVNEEDGE GDAPDATTQP QKRKRLWGLV VCHNTTPRFV PFPLRYACEF LAQVFAIHVN KEVELDNQMV EKNILRTQTL LCDMLMRDAP LGIVSQSPNI MDLVKCDGAA LLYKDKIWKL GTTPSEFHLQ EIASWLCEYH MDSTGLSTDS LHDAGFPRAL SLGDSVCGMA AVRISSKDMI FWFRSHTAGE VRWGGAKHDP DDRDDARRMH PRSSFKAFLE VVKTRSLPWK DYEMDAIHSL QLILRNAFKD SETTDVNTKV IYSKLNDLKI DGIQELEAVT SEMVRLIETA TVPILAVDSD GLVNGWNTKI AELTGLSVDE AIGKHFLTLV EDSSVEIVKR MLENALEGTE EQNVQFEIKT HLSRADAGPI SLVVNACASR DLHENVVGVC FVAHDLTGQK TVMDKFTRIE GDYKAIIQNP NPLIPPIFGT DEFGWCTEWN PAMSKLTGLK REEVIDKMLL GEVFGTQKSC CRLKNQEAFV NLGIVLNNAV TSQDPEKVSF AFFTRGGKYV ECLLCVSKKL DREGVVTGVF CFLQLASHEL QQALHVQRLA ERTAVKRLKA LAYIKRQIRN PLSGIMFTRK MIEGTELGPE QRRILQTSAL CQKQLSKILD DSDLESIIEG CLDLEMKEFT LNEVLTASTS QVMMKSNGKS VRITNETGEE VMSDTLYGDS IRLQQVLADF MLMAVNFTPS GGQLTVSASL RKDQLGRSVH LANLEIRLTH TGAGIPEFLL NQMFGTEEDV SEEGLSLMVS RKLVKLMNGD VQYLRQAGKS SFIITAELAA ANK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 299 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 193.0 K / Max: 193.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.05 mrad |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 6390 / Average exposure time: 1.0 sec. / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)