+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human ABCA4 structure in complex with AMP-PNP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / importer / membrane protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationrod photoreceptor disc membrane / flippase activity / N-retinylidene-phosphatidylethanolamine flippase activity / Defective visual phototransduction due to ABCA4 loss of function / all-trans retinal binding / retinol transmembrane transporter activity / phospholipid transfer to membrane / phospholipid transporter activity / 11-cis retinal binding / ATPase-coupled intramembrane lipid transporter activity ...rod photoreceptor disc membrane / flippase activity / N-retinylidene-phosphatidylethanolamine flippase activity / Defective visual phototransduction due to ABCA4 loss of function / all-trans retinal binding / retinol transmembrane transporter activity / phospholipid transfer to membrane / phospholipid transporter activity / 11-cis retinal binding / ATPase-coupled intramembrane lipid transporter activity / retinal metabolic process / phosphatidylethanolamine flippase activity / retinoid binding / photoreceptor cell maintenance / P-type phospholipid transporter / phospholipid translocation / lipid transport / The canonical retinoid cycle in rods (twilight vision) / phototransduction, visible light / ATPase-coupled transmembrane transporter activity / photoreceptor outer segment / ABC-type transporter activity / retinoid metabolic process / visual perception / ABC-family proteins mediated transport / transmembrane transport / photoreceptor disc membrane / cytoplasmic vesicle / intracellular membrane-bounded organelle / GTPase activity / endoplasmic reticulum / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Scortecci JF / Van Petegem F / Molday RS | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: Structural and functional characterization of the nucleotide-binding domains of ABCA4 and their role in Stargardt disease. Authors: Jessica Fernandes Scortecci / Fabian A Garces / Jai K Mahto / Laurie L Molday / Filip Van Petegem / Robert S Molday /  Abstract: ABCA4 is an ATP-binding cassette (ABC) transporter that prevents the buildup of toxic retinoid compounds by facilitating the transport of N-retinylidene-phosphatidylethanolamine across membranes of ...ABCA4 is an ATP-binding cassette (ABC) transporter that prevents the buildup of toxic retinoid compounds by facilitating the transport of N-retinylidene-phosphatidylethanolamine across membranes of rod and cone photoreceptor cells. Over 1500 missense mutations in ABCA4, many in the nucleotide-binding domains (NBDs), have been genetically linked to Stargardt disease. Here, we show by cryo-EM that ABCA4 is converted from an open outward conformation to a closed conformation upon the binding of adenylyl-imidodiphosphate. Structural information and biochemical studies were used to further define the role of the NBDs in the functional properties of ABCA4 and the mechanisms by which mutations lead to the loss in activity. We show that ATPase activity in both NBDs is required for the functional activity of ABCA4. Mutations in Walker A asparagine residues cause a severe reduction in substrate-activated ATPase activity due to the loss in polar interactions with residues within the D-loops of the opposing NBD. The structural basis for how disease mutations in other NBD residues, including the R1108C, R2077W, R2107H, and L2027F, affect the structure and function of ABCA4 is described. Collectively, our studies provide insight into the structure and function of ABCA4 and mechanisms underlying Stargardt disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28864.map.gz emd_28864.map.gz | 89.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28864-v30.xml emd-28864-v30.xml emd-28864.xml emd-28864.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28864.png emd_28864.png | 13.8 KB | ||
| Filedesc metadata |  emd-28864.cif.gz emd-28864.cif.gz | 7.3 KB | ||
| Others |  emd_28864_half_map_1.map.gz emd_28864_half_map_1.map.gz emd_28864_half_map_2.map.gz emd_28864_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28864 http://ftp.pdbj.org/pub/emdb/structures/EMD-28864 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28864 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28864 | HTTPS FTP |
-Related structure data
| Related structure data |  8f5bMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28864.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28864.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04903 Å | ||||||||||||||||||||||||||||||||||||
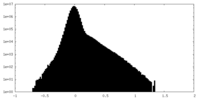
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28864_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
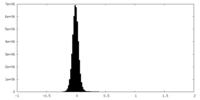
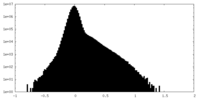
| Density Histograms |
-Half map: #1
| File | emd_28864_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
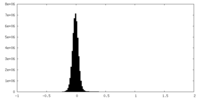
| Density Histograms |
- Sample components
Sample components
-Entire : ABCA4 structure in complex with AMP-PNP
| Entire | Name: ABCA4 structure in complex with AMP-PNP |
|---|---|
| Components |
|
-Supramolecule #1: ABCA4 structure in complex with AMP-PNP
| Supramolecule | Name: ABCA4 structure in complex with AMP-PNP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Recombinantly expressed human ABCA4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Retinal-specific phospholipid-transporting ATPase ABCA4
| Macromolecule | Name: Retinal-specific phospholipid-transporting ATPase ABCA4 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: P-type phospholipid transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 256.202531 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGFVRQIQLL LWKNWTLRKR QKIRFVVELV WPLSLFLVLI WLRNANPLYS HHECHFPNKA MPSAGMLPWL QGIFCNVNNP CFQSPTPGE SPGIVSNYNN SILARVYRDF QELLMNAPES QHLGRIWTEL HILSQFMDTL RTHPERIAGR GIRIRDILKD E ETLTLFLI ...String: MGFVRQIQLL LWKNWTLRKR QKIRFVVELV WPLSLFLVLI WLRNANPLYS HHECHFPNKA MPSAGMLPWL QGIFCNVNNP CFQSPTPGE SPGIVSNYNN SILARVYRDF QELLMNAPES QHLGRIWTEL HILSQFMDTL RTHPERIAGR GIRIRDILKD E ETLTLFLI KNIGLSDSVV YLLINSQVRP EQFAHGVPDL ALKDIACSEA LLERFIIFSQ RRGAKTVRYA LCSLSQGTLQ WI EDTLYAN VDFFKLFRVL PTLLDSRSQG INLRSWGGIL SDMSPRIQEF IHRPSMQDLL WVTRPLMQNG GPETFTKLMG ILS DLLCGY PEGGGSRVLS FNWYEDNNYK AFLGIDSTRK DPIYSYDRRT TSFCNALIQS LESNPLTKIA WRAAKPLLMG KILY TPDSP AARRILKNAN STFEELEHVR KLVKAWEEVG PQIWYFFDNS TQMNMIRDTL GNPTVKDFLN RQLGEEGITA EAILN FLYK GPRESQADDM ANFDWRDIFN ITDRTLRLVN QYLECLVLDK FESYNDETQL TQRALSLLEE NMFWAGVVFP DMYPWT SSL PPHVKYKIRM DIDVVEKTNK IKDRYWDSGP RADPVEDFRY IWGGFAYLQD MVEQGITRSQ VQAEAPVGIY LQQMPYP CF VDDSFMIILN RCFPIFMVLA WIYSVSMTVK SIVLEKELRL KETLKNQGVS NAVIWCTWFL DSFSIMSMSI FLLTIFIM H GRILHYSDPF ILFLFLLAFS TATIMLCFLL STFFSKASLA AACSGVIYFT LYLPHILCFA WQDRMTAELK KAVSLLSPV AFGFGTEYLV RFEEQGLGLQ WSNIGNSPTE GDEFSFLLSM QMMLLDAAVY GLLAWYLDQV FPGDYGTPLP WYFLLQESYW LGGEGCSTR EERALEKTEP LTEETEDPEH PEGIHDSFFE REHPGWVPGV CVKNLVKIFE PCGRPAVDRL NITFYENQIT A FLGHNGAG KTTTLSILTG LLPPTSGTVL VGGRDIETSL DAVRQSLGMC PQHNILFHHL TVAEHMLFYA QLKGKSQEEA QL EMEAMLE DTGLHHKRNE EAQDLSGGMQ RKLSVAIAFV GDAKVVILDE PTSGVDPYSR RSIWDLLLKY RSGRTIIMST HHM DEADLL GDRIAIIAQG RLYCSGTPLF LKNCFGTGLY LTLVRKMKNI QSQRKGSEGT CSCSSKGFST TCPAHVDDLT PEQV LDGDV NELMDVVLHH VPEAKLVECI GQELIFLLPN KNFKHRAYAS LFRELEETLA DLGLSSFGIS DTPLEEIFLK VTEDS DSGP LFAGGAQQKR ENVNPRHPCL GPREKAGQTP QDSNVCSPGA PAAHPEGQPP PEPECPGPQL NTGTQLVLQH VQALLV KRF QHTIRSHKDF LAQIVLPATF VFLALMLSIV IPPFGEYPAL TLHPWIYGQQ YTFFSMDEPG SEQFTVLADV LLNKPGF GN RCLKEGWLPE YPCGNSTPWK TPSVSPNITQ LFQKQKWTQV NPSPSCRCST REKLTMLPEC PEGAGGLPPP QRTQRSTE I LQDLTDRNIS DFLVKTYPAL IRSSLKSKFW VNEQRYGGIS IGGKLPVVPI TGEALVGFLS DLGRIMNVSG GPITREASK EIPDFLKHLE TEDNIKVWFN NKGWHALVSF LNVAHNAILR ASLPKDRSPE EYGITVISQP LNLTKEQLSE ITVLTTSVDA VVAICVIFS MSFVPASFVL YLIQERVNKS KHLQFISGVS PTTYWVTNFL WDIMNYSVSA GLVVGIFIGF QKKAYTSPEN L PALVALLL LYGWAVIPMM YPASFLFDVP STAYVALSCA NLFIGINSSA ITFILELFEN NRTLLRFNAV LRKLLIVFPH FC LGRGLID LALSQAVTDV YARFGEEHSA NPFHWDLIGK NLFAMVVEGV VYFLLTLLVQ RHFFLSQWIA EPTKEPIVDE DDD VAEERQ RIITGGNKTD ILRLHELTKI YPGTSSPAVD RLCVGVRPGE CFGLLGVNGA GKTTTFKMLT GDTTVTSGDA TVAG KSILT NISEVHQNMG YCPQFDAIDE LLTGREHLYL YARLRGVPAE EIEKVANWSI KSLGLTVYAD CLAGTYSGGN KRKLS TAIA LIGCPPLVLL DEPTTGMDPQ ARRMLWNVIV SIIREGRAVV LTSHSMEECE ALCTRLAIMV KGAFRCMGTI QHLKSK FGD GYIVTMKIKS PKDDLLPDLN PVEQFFQGNF PGSVQRERHY NMLQFQVSSS SLARIFQLLL SHKDSLLIEE YSVTQTT LD QVFVNFAKQQ TESHDLPLHP RAAGASRQAQ D UniProtKB: Retinal-specific phospholipid-transporting ATPase ABCA4 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)