+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Gentamicin bound aminoglycoside efflux pump AcrD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | aminoglycoside efflux pump / AcrD / E.coli / MEMBRANE PROTEIN | |||||||||
| Function / homology | Hydrophobe/amphiphile efflux-1 HAE1 / Multidrug efflux transporter AcrB TolC docking domain, DN/DC subdomains / Acriflavin resistance protein / AcrB/AcrD/AcrF family / efflux transmembrane transporter activity / xenobiotic transmembrane transporter activity / response to toxic substance / plasma membrane / Efflux pump membrane transporter Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
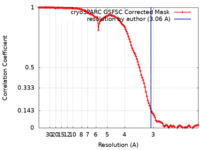
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Zhang Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2023 Journal: mBio / Year: 2023Title: Cryo-EM Structures of AcrD Illuminate a Mechanism for Capturing Aminoglycosides from Its Central Cavity. Authors: Zhemin Zhang / Christopher E Morgan / Meng Cui / Edward W Yu /  Abstract: The Escherichia coli acriflavine resistance protein D (AcrD) is an efflux pump that belongs to the resistance-nodulation-cell division (RND) superfamily. Its primary function is to provide resistance ...The Escherichia coli acriflavine resistance protein D (AcrD) is an efflux pump that belongs to the resistance-nodulation-cell division (RND) superfamily. Its primary function is to provide resistance to aminoglycoside-based drugs by actively extruding these noxious compounds out of E. coli cells. AcrD can also mediate resistance to a limited range of other amphiphilic agents, including bile acids, novobiocin, and fusidic acids. As there is no structural information available for any aminoglycoside-specific RND pump, here we describe cryo-electron microscopy (cryo-EM) structures of AcrD in the absence and presence of bound gentamicin. These structures provide new information about the RND superfamily of efflux pumps, specifically, that three negatively charged residues central to the aminoglycoside-binding site are located within the ceiling of the central cavity of the AcrD trimer. Thus, it is likely that AcrD is capable of picking up aminoglycosides via this central cavity. Through the combination of cryo-EM structural determination, mutagenesis analysis, and molecular simulation, we show that charged residues are critically important for this pump to shuttle drugs directly from the central cavity to the funnel of the AcrD trimer for extrusion. Here, we report cryo-EM structures of the AcrD aminoglycoside efflux pump in the absence and presence of bound gentamicin, posing the possibility that this pump is capable of capturing aminoglycosides from the central cavity of the AcrD trimer. The results indicate that AcrD utilizes charged residues to bind and export drugs, mediating resistance to these antibiotics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28857.map.gz emd_28857.map.gz | 157.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28857-v30.xml emd-28857-v30.xml emd-28857.xml emd-28857.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28857_fsc.xml emd_28857_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28857.png emd_28857.png | 172.7 KB | ||
| Filedesc metadata |  emd-28857.cif.gz emd-28857.cif.gz | 7.5 KB | ||
| Others |  emd_28857_additional_1.map.gz emd_28857_additional_1.map.gz emd_28857_half_map_1.map.gz emd_28857_half_map_1.map.gz emd_28857_half_map_2.map.gz emd_28857_half_map_2.map.gz | 83.9 MB 154.2 MB 154.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28857 http://ftp.pdbj.org/pub/emdb/structures/EMD-28857 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28857 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28857 | HTTPS FTP |
-Validation report
| Summary document |  emd_28857_validation.pdf.gz emd_28857_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28857_full_validation.pdf.gz emd_28857_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_28857_validation.xml.gz emd_28857_validation.xml.gz | 20.7 KB | Display | |
| Data in CIF |  emd_28857_validation.cif.gz emd_28857_validation.cif.gz | 26.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28857 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28857 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28857 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28857 | HTTPS FTP |
-Related structure data
| Related structure data |  8f4rMC  8f3eC  8f4nC  8f56C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28857.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28857.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
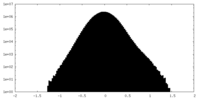
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_28857_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
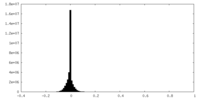
| Density Histograms |
-Half map: #2
| File | emd_28857_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
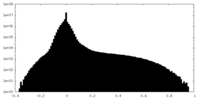
| Density Histograms |
-Half map: #1
| File | emd_28857_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
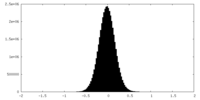
| Density Histograms |
- Sample components
Sample components
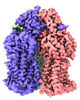
-Entire : AcrD
| Entire | Name: AcrD |
|---|---|
| Components |
|
-Supramolecule #1: AcrD
| Supramolecule | Name: AcrD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Efflux pump membrane transporter
| Macromolecule | Name: Efflux pump membrane transporter / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 113.139891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANFFIDRPI FAWVLAILLC LTGTLAIFSL PVEQYPDLAP PNVRVTANYP GASAQTLENT VTQVIEQNMT GLDNLMYMSS QSSGTGQAS VTLSFKAGTD PDEAVQQVQN QLQSAMRKLP QAVQNQGVTV RKTGDTNILT IAFVSTDGSM DKQDIADYVA S NIQDPLSR ...String: MANFFIDRPI FAWVLAILLC LTGTLAIFSL PVEQYPDLAP PNVRVTANYP GASAQTLENT VTQVIEQNMT GLDNLMYMSS QSSGTGQAS VTLSFKAGTD PDEAVQQVQN QLQSAMRKLP QAVQNQGVTV RKTGDTNILT IAFVSTDGSM DKQDIADYVA S NIQDPLSR VNGVGDIDAY GSQYSMRIWL DPAKLNSFQM TAKDVTDAIE SQNAQIAVGQ LGGTPSVDKQ ALNATINAQS LL QTPEQFR DITLRVNQDG SEVRLGDVAT VEMGAEKYDY LSRFNGKPAS GLGVKLASGA NEMATAELVL NRLDELAQYF PHG LEYKVA YETTSFVKAS IEDVVKTLLE AIALVFLVMY LFLQNFRATL IPTIAVPVVL MGTFSVLYAF GYSVNTLTMF AMVL AIGLL VDDAIVVVEN VERIMSEEGL TPREATRKSM GQIQGALVGI AMVLSAVFVP MAFFGGTTGA IYRQFSITIV AAMVL SVLV AMILTPALCA TLLKPLKKGE HHGQKGFFAW FNQMFNRNAE RYEKGVAKIL HRSLRWIVIY VLLLGGMVFL FLRLPT SFL PLEDRGMFTT SVQLPSGSTQ QQTLKVVEQI EKYYFTHEKD NIMSVFATVG SGPGGNGQNV ARMFIRLKDW SERDSKT GT SFAIIERATK AFNQIKEARV IASSPPAISG LGSSAGFDME LQDHAGAGHD ALMAARNQLL ALAAENPELT RVRHNGLD D SPQLQIDIDQ RKAQALGVAI DDINDTLQTA WGSSYVNDFM DRGRVKKVYV QAAAPYRMLP DDINLWYVRN KDGGMVPFS AFATSRWETG SPRLERYNGY SAVEIVGEAA PGVSTGTAMD IMESLVKQLP NGFGLEWTAM SYQERLSGAQ APALYAISLL VVFLCLAAL YESWSVPFSV MLVVPLGVIG ALLATWMRGL ENDVYFQVGL LTVIGLSAKN AILIVEFANE MNQKGHDLFE A TLHACRQR LRPILMTSLA FIFGVLPMAT STGAGSGGQH AVGTGVMGGM ISATILAIYF VPLFFVLVRR RFPLKPRPE UniProtKB: Efflux pump membrane transporter |
-Macromolecule #2: (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMI...
| Macromolecule | Name: (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL type: ligand / ID: 2 / Number of copies: 1 / Formula: LLL |
|---|---|
| Molecular weight | Theoretical: 449.542 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 35.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)