[English] 日本語
 Yorodumi
Yorodumi- EMDB-28826: Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
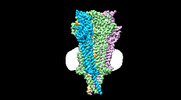
| Title | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, pore-blocked state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | acetylcholine receptor / nicotinic receptor / Torpedo / Cys-loop receptor / ion channel / muscle-type nicotinic receptor / TRANSPORT PROTEIN-INHIBITOR complex / Neuromuscular Blocker / Inhibitor | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine-gated monoatomic cation-selective channel activity / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / transmembrane signaling receptor activity / postsynaptic membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
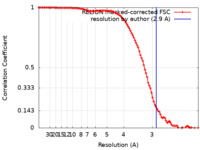
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Goswami U / Rahman MM / Teng J / Hibbs RE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural interplay of anesthetics and paralytics on muscle nicotinic receptors. Authors: Umang Goswami / Md Mahfuzur Rahman / Jinfeng Teng / Ryan E Hibbs /  Abstract: General anesthetics and neuromuscular blockers are used together during surgery to stabilize patients in an unconscious state. Anesthetics act mainly by potentiating inhibitory ion channels and ...General anesthetics and neuromuscular blockers are used together during surgery to stabilize patients in an unconscious state. Anesthetics act mainly by potentiating inhibitory ion channels and inhibiting excitatory ion channels, with the net effect of dampening nervous system excitability. Neuromuscular blockers act by antagonizing nicotinic acetylcholine receptors at the motor endplate; these excitatory ligand-gated ion channels are also inhibited by general anesthetics. The mechanisms by which anesthetics and neuromuscular blockers inhibit nicotinic receptors are poorly understood but underlie safe and effective surgeries. Here we took a direct structural approach to define how a commonly used anesthetic and two neuromuscular blockers act on a muscle-type nicotinic receptor. We discover that the intravenous anesthetic etomidate binds at an intrasubunit site in the transmembrane domain and stabilizes a non-conducting, desensitized-like state of the channel. The depolarizing neuromuscular blocker succinylcholine also stabilizes a desensitized channel but does so through binding to the classical neurotransmitter site. Rocuronium binds in this same neurotransmitter site but locks the receptor in a resting, non-conducting state. Together, this study reveals a structural mechanism for how general anesthetics work on excitatory nicotinic receptors and further rationalizes clinical observations in how general anesthetics and neuromuscular blockers interact. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28826.map.gz emd_28826.map.gz | 71.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28826-v30.xml emd-28826-v30.xml emd-28826.xml emd-28826.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28826_fsc.xml emd_28826_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_28826.png emd_28826.png | 40.2 KB | ||
| Filedesc metadata |  emd-28826.cif.gz emd-28826.cif.gz | 7.5 KB | ||
| Others |  emd_28826_half_map_1.map.gz emd_28826_half_map_1.map.gz emd_28826_half_map_2.map.gz emd_28826_half_map_2.map.gz | 71.3 MB 71.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28826 http://ftp.pdbj.org/pub/emdb/structures/EMD-28826 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28826 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28826 | HTTPS FTP |
-Related structure data
| Related structure data |  8f2sMC  8eskC  8f6yC  8f6zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28826.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28826.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0694 Å | ||||||||||||||||||||||||||||||||||||
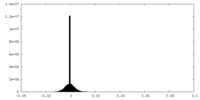
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28826_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
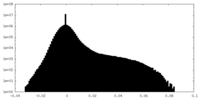
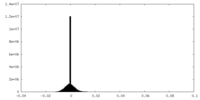
| Density Histograms |
-Half map: #1
| File | emd_28826_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
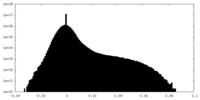
| Density Histograms |
- Sample components
Sample components
+Entire : Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in ...
+Supramolecule #1: Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in ...
+Macromolecule #1: Acetylcholine receptor subunit alpha
+Macromolecule #2: Acetylcholine receptor subunit delta
+Macromolecule #3: Acetylcholine receptor subunit beta
+Macromolecule #4: Acetylcholine receptor subunit gamma
+Macromolecule #9: rocuronium
+Macromolecule #10: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
+Macromolecule #11: CHOLESTEROL
+Macromolecule #12: 2-acetamido-2-deoxy-beta-D-glucopyranose
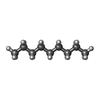
+Macromolecule #13: nonane
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.5 µm / Nominal defocus min: 2.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)