[English] 日本語
 Yorodumi
Yorodumi- EMDB-28818: Structure of yeast F1-ATPase determined with 100 micromolar cruen... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of yeast F1-ATPase determined with 100 micromolar cruentaren A | ||||||||||||||||||
 Map data Map data | Sharpened map. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | F1-ATPase / ATP Synthase / cruentaren A / drug development / HYDROLASE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information: / : / : / : / proton motive force-driven mitochondrial ATP synthesis / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / ATP hydrolysis activity / ATP binding Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
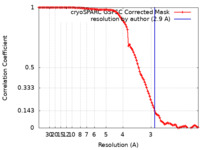
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||||||||
 Authors Authors | Guo H / Rubinstein JL | ||||||||||||||||||
| Funding support |  Canada, Canada,  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Chemistry / Year: 2023 Journal: Chemistry / Year: 2023Title: CryoEM Structure with ATP Synthase Enables Late-Stage Diversification of Cruentaren A. Authors: Xiaozheng Dou / Hui Guo / Terin D'Amico / Leah Abdallah / Chitra Subramanian / Bhargav A Patel / Mark Cohen / John L Rubinstein / Brian S J Blagg /   Abstract: Cruentaren A is a natural product that exhibits potent antiproliferative activity against various cancer cell lines, yet its binding site within ATP synthase remained unknown, thus limiting the ...Cruentaren A is a natural product that exhibits potent antiproliferative activity against various cancer cell lines, yet its binding site within ATP synthase remained unknown, thus limiting the development of improved analogues as anticancer agents. Herein, we report the cryogenic electron microscopy (cryoEM) structure of cruentaren A bound to ATP synthase, which allowed the design of new inhibitors through semisynthetic modification. Examples of cruentaren A derivatives include a trans-alkene isomer, which was found to exhibit similar activity to cruentaren A against three cancer cell lines as well as several other analogues that retained potent inhibitory activity. Together, these studies provide a foundation for the generation of cruentaren A derivatives as potential therapeutics for the treatment of cancer. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28818.map.gz emd_28818.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28818-v30.xml emd-28818-v30.xml emd-28818.xml emd-28818.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28818_fsc.xml emd_28818_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_28818.png emd_28818.png | 83.1 KB | ||
| Masks |  emd_28818_msk_1.map emd_28818_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28818.cif.gz emd-28818.cif.gz | 7.3 KB | ||
| Others |  emd_28818_additional_1.map.gz emd_28818_additional_1.map.gz emd_28818_half_map_1.map.gz emd_28818_half_map_1.map.gz emd_28818_half_map_2.map.gz emd_28818_half_map_2.map.gz | 62.5 MB 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28818 http://ftp.pdbj.org/pub/emdb/structures/EMD-28818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28818 | HTTPS FTP |
-Validation report
| Summary document |  emd_28818_validation.pdf.gz emd_28818_validation.pdf.gz | 900.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28818_full_validation.pdf.gz emd_28818_full_validation.pdf.gz | 900.3 KB | Display | |
| Data in XML |  emd_28818_validation.xml.gz emd_28818_validation.xml.gz | 18.7 KB | Display | |
| Data in CIF |  emd_28818_validation.cif.gz emd_28818_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28818 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28818 | HTTPS FTP |
-Related structure data
| Related structure data |  8f2kMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28818.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28818.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04622 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28818_msk_1.map emd_28818_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
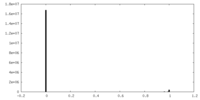
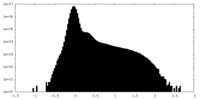
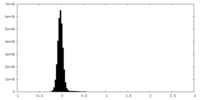
| Density Histograms |
-Additional map: Unsharpened map.
| File | emd_28818_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
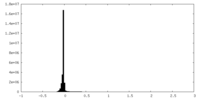
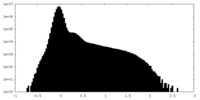
| Density Histograms |
-Half map: Half map 1.
| File | emd_28818_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
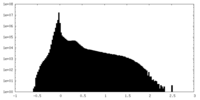
| Density Histograms |
-Half map: Half map 2.
| File | emd_28818_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
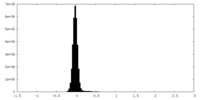
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast F1-ATPase with 100 micromolar cruentaren A
| Entire | Name: Yeast F1-ATPase with 100 micromolar cruentaren A |
|---|---|
| Components |
|
-Supramolecule #1: Yeast F1-ATPase with 100 micromolar cruentaren A
| Supramolecule | Name: Yeast F1-ATPase with 100 micromolar cruentaren A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: ATP synthase subunit alpha
| Macromolecule | Name: ATP synthase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.376539 KDa |
| Sequence | String: NLNETGRVLA VGDGIARVFG LNNIQAEELV EFSSGVKGMA LNLEPGQVGI VLFGSDRLVK EGELVKRTGN IVDVPVGPGL LGRVVDALG NPIDGKGPID AAGRSRAQVK APGILPRRSV HEPVQTGLKA VDALVPIGRG QRELIIGDRQ TGKTAVALDT I LNQKRWNN ...String: NLNETGRVLA VGDGIARVFG LNNIQAEELV EFSSGVKGMA LNLEPGQVGI VLFGSDRLVK EGELVKRTGN IVDVPVGPGL LGRVVDALG NPIDGKGPID AAGRSRAQVK APGILPRRSV HEPVQTGLKA VDALVPIGRG QRELIIGDRQ TGKTAVALDT I LNQKRWNN GSDESKKLYC VYVAVGQKRS TVAQLVQTLE QHDAMKYSII VAATASEAAP LQYLAPFTAA SIGEWFRDNG KH ALIVYDD LSKQAVAYRQ LSLLLRRPPG REAYPGDVFY LHSRLLERAA KLSEKEGSGS LTALPVIETQ GGDVSAYIPT NVI SITDGQ IFLEAELFYK GIRPAINVGL SVSRVGSAAQ VKALKQVAGS LKLFLAQYRE VAAFAQFGSD LDASTKQTLV RGER LTQLL KQNQYSPLAT EEQVPLIYAG VNGHLDGIEL SRIGEFESSF LSYLKSNHNE LLTEIREKGE LSKELLASLK SATES FVAT F UniProtKB: ATP synthase subunit alpha |
-Macromolecule #2: ATP synthase subunit beta
| Macromolecule | Name: ATP synthase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: H+-transporting two-sector ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.379277 KDa |
| Sequence | String: PITGKVTAVI GAIVDVHFEQ SELPAILNAL EIKTPQGKLV LEVAQHLGEN TVRTIAMDGT EGLVRGEKVL DTGGPISVPV GRETLGRII NVIGEPIDER GPIKSKLRKP IHADPPSFAE QSTSAEILET GIKVVDLLAP YARGGKIGLF GGAGVGKTVF I QELINNIA ...String: PITGKVTAVI GAIVDVHFEQ SELPAILNAL EIKTPQGKLV LEVAQHLGEN TVRTIAMDGT EGLVRGEKVL DTGGPISVPV GRETLGRII NVIGEPIDER GPIKSKLRKP IHADPPSFAE QSTSAEILET GIKVVDLLAP YARGGKIGLF GGAGVGKTVF I QELINNIA KAHGGFSVFT GVGERTREGN DLYREMKETG VINLEGESKV ALVFGQMNEP PGARARVALT GLTIAEYFRD EE GQDVLLF IDNIFRFTQA GSEVSALLGR IPSAVGYQPT LATDMGLLQE RITTTKKGSV TSVQAVYVPA DDLTDPAPAT TFA HLDATT VLSRGISELG IYPAVDPLDS KSRLLDAAVV GQEHYDVASK VQETLQTYKS LQDIIAILGM DELSEQDKLT VERA RKIQR FLSQPFAVAE VFTGIPGKLV RLKDTVASFK AVLEGKYDNI PEHAFYMVGG IEDVVAKAEK LAAE UniProtKB: ATP synthase subunit beta |
-Macromolecule #3: ATP synthase subunit gamma
| Macromolecule | Name: ATP synthase subunit gamma / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.399875 KDa |
| Sequence | String: ATLKEVEMRL KSIKNIEKIT KTMKIVASTR LSKAEKAKIS AKKMDEAEQL FYKNAETKNL DVEATETGAP KELIVAITSD KGLCGSIHS QLAKAVRRHL NDQPNADIVT IGDKIKMQLL RTHPNNIKLS INGIGKDAPT FQESALIADK LLSVMKAGTY P KISIFYND ...String: ATLKEVEMRL KSIKNIEKIT KTMKIVASTR LSKAEKAKIS AKKMDEAEQL FYKNAETKNL DVEATETGAP KELIVAITSD KGLCGSIHS QLAKAVRRHL NDQPNADIVT IGDKIKMQLL RTHPNNIKLS INGIGKDAPT FQESALIADK LLSVMKAGTY P KISIFYND PVSSLSFEPS EKPIFNAKTI EQSPSFGKFE IDTDANVPRD LFEYTLANQM LTAMAQGYAA EISARRNAMD NA SKNAGDM INRYSILYNR TRQAVITNEL VDIITGAS UniProtKB: ATP synthase subunit gamma |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 5 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: Cruentaren A
| Macromolecule | Name: Cruentaren A / type: ligand / ID: 6 / Number of copies: 2 / Formula: XBC |
|---|---|
| Molecular weight | Theoretical: 589.76 Da |
| Chemical component information | 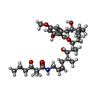 ChemComp-XBC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 35 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 5814 / Average exposure time: 8.4 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 133815 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)