[English] 日本語
 Yorodumi
Yorodumi- EMDB-28815: Cryo-EM of the S. cerevisiae chromatin remodeler Yta7 hexamer bou... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to nucleosome | |||||||||
 Map data Map data | EM map un sharpen and vop gaussian processed | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ ATPase / chromatin remodeler / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP-dependent histone chaperone activity / positive regulation of invasive growth in response to glucose limitation / CENP-A containing chromatin assembly / ATP-dependent chromatin remodeler activity / nucleosome disassembly / chromosome, centromeric region / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / transcription initiation-coupled chromatin remodeling / DNA-templated DNA replication / nucleosome assembly ...ATP-dependent histone chaperone activity / positive regulation of invasive growth in response to glucose limitation / CENP-A containing chromatin assembly / ATP-dependent chromatin remodeler activity / nucleosome disassembly / chromosome, centromeric region / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / transcription initiation-coupled chromatin remodeling / DNA-templated DNA replication / nucleosome assembly / chromosome / chromatin organization / histone binding / chromatin remodeling / chromatin binding / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / ATP binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
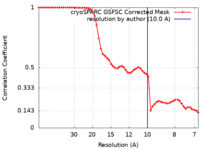
| Method | single particle reconstruction / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Wang F / Feng X / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2023 Journal: J Biol Chem / Year: 2023Title: The Saccharomyces cerevisiae Yta7 ATPase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly. Authors: Feng Wang / Xiang Feng / Qing He / Hua Li / Huilin Li /  Abstract: The Saccharomyces cerevisiae Yta7 is a chromatin remodeler harboring a histone-interacting bromodomain (BRD) and two AAA+ modules. It is not well understood how Yta7 recognizes the histone H3 tail to ...The Saccharomyces cerevisiae Yta7 is a chromatin remodeler harboring a histone-interacting bromodomain (BRD) and two AAA+ modules. It is not well understood how Yta7 recognizes the histone H3 tail to promote nucleosome disassembly for DNA replication or RNA transcription. By cryo-EM analysis, here we show that Yta7 assembles a three-tiered hexamer with a top BRD tier, a middle AAA1 tier, and a bottom AAA2 tier. Unexpectedly, the Yta7 BRD stabilizes a four-stranded β-helix, termed BRD-interacting motif (BIM), of the largely disordered N-terminal region. The BIM motif is unique to the baker's yeast, and we show both BRD and BIM contribute to nucleosome recognition. We found that Yta7 binds both acetylated and nonacetylated H3 peptides but with a higher affinity for the unmodified peptide. This property is consistent with the absence of key residues of canonical BRDs involved in acetylated peptide recognition and the role of Yta7 in general nucleosome remodeling. Interestingly, the BRD tier exists in a spiral and a flat-ring form on top of the Yta7 AAA+ hexamer. The spiral is likely in a nucleosome-searching mode because the bottom BRD blocks the entry to the AAA+ chamber. The flat ring may be in a nucleosome disassembly state because the entry is unblocked and the H3 peptide has entered the AAA+ chamber and is stabilized by the AAA1 pore loops 1 and 2. Indeed, we show that the BRD tier is a flat ring when bound to the nucleosome. Overall, our study sheds light on the nucleosome disassembly by Yta7. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28815.map.gz emd_28815.map.gz | 13.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28815-v30.xml emd-28815-v30.xml emd-28815.xml emd-28815.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28815_fsc.xml emd_28815_fsc.xml | 5.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_28815.png emd_28815.png | 22.9 KB | ||
| Others |  emd_28815_half_map_1.map.gz emd_28815_half_map_1.map.gz emd_28815_half_map_2.map.gz emd_28815_half_map_2.map.gz | 14.4 MB 14.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28815 http://ftp.pdbj.org/pub/emdb/structures/EMD-28815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28815 | HTTPS FTP |
-Validation report
| Summary document |  emd_28815_validation.pdf.gz emd_28815_validation.pdf.gz | 644.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28815_full_validation.pdf.gz emd_28815_full_validation.pdf.gz | 643.7 KB | Display | |
| Data in XML |  emd_28815_validation.xml.gz emd_28815_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_28815_validation.cif.gz emd_28815_validation.cif.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28815 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28815 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28815 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28815 | HTTPS FTP |
-Related structure data
| Related structure data |  7uqiC  7uqjC  7uqkC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28815.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28815.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map un sharpen and vop gaussian processed | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.312 Å | ||||||||||||||||||||||||||||||||||||
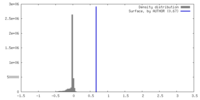
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map
| File | emd_28815_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
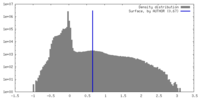
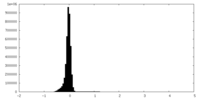
| Density Histograms |
-Half map: Half map
| File | emd_28815_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
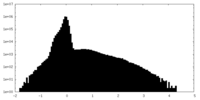
| Density Histograms |
- Sample components
Sample components
-Entire : Yta7 and nucleosome complex
| Entire | Name: Yta7 and nucleosome complex |
|---|---|
| Components |
|
-Supramolecule #1: Yta7 and nucleosome complex
| Supramolecule | Name: Yta7 and nucleosome complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Gra-fix fractions that contain Yta7 and nucleosome |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 930 KDa |
-Macromolecule #1: Yta7
| Macromolecule | Name: Yta7 / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MARNLRNRRG SDVEDASNAK VGYETQIKDE NGIIHTTTRS LRKINYAEIE KVFDFLEDDQ VMDKDETPVD VTSDEHHNNN QKGDDEDDDV DLVSPHENAR TNEELTNERN LRKRKAHDPE EDDESFHEED VDDDEEEEEA DEFEDEYLDE DSKDNNRRRR AADRKFVVPD ...String: MARNLRNRRG SDVEDASNAK VGYETQIKDE NGIIHTTTRS LRKINYAEIE KVFDFLEDDQ VMDKDETPVD VTSDEHHNNN QKGDDEDDDV DLVSPHENAR TNEELTNERN LRKRKAHDPE EDDESFHEED VDDDEEEEEA DEFEDEYLDE DSKDNNRRRR AADRKFVVPD PDDDEEYDED DEEGDRISHS ASSKRLKRAN SRRTRSSRHP ETPPPVRRAL RSRTRHSRTS NEENDDENDN SRNEALTLAD EIRELQEDSP IREKRFLRER TKPVNYKLPP PLTASNAEEF IDKNNNALSF HNPSPARRGR GGWNASQNSG PTRRLFPTGG PFGGNDVTTI FGKNTNFYNQ VPSAFSDNNN NKLILDSDSS DDEILPLGVT PKTKKENTQK KKKKKPEIAD LDPLGVDMNV NFDDIGGLDN YIDQLKEMVA LPLLYPELYQ NFNITPPRGV LFHGPPGTGK TLMARALAAS CSSDERKITF FMRKGADILS KWVGEAERQL RLLFEEAKKH QPSIIFFDEI DGLAPVRSSK QEQIHASIVS TLLALMDGMD NRGQVIVIGA TNRPDAVDPA LRRPGRFDRE FYFPLPDVKA RFKILQIQTR KWSSPLSTNF IDKLAFLTKG YGGADLRSLC TEAALISIQR SFPQIYRSND KLLVDPSKIK VKVSDFMLAL KKIVPSSARS TGSSPQPLPE LIKPLLADQL NNLKNKLDYM LNIKDTTFQR NTSLLQNFID YEEYSGEEEE HDKYGGNEDT SSFRSYEFFE SMAESQICKP RLLINGPKGN GQQYVGAAIL NYLEEFNVQN LDLASLVSES SRTIEAAVVQ SFMEAKKRQP SVVFIPNLDI WINTIPENVI LVLSGLFRSL QSNEKILLLC LAENLDISEV KNGILSDFAF DKNIFQLHKP SKENITRYFS NLIELLKTKP SDIPMKKRRV KPLPELQKVT SNAAPTNFDE NGEPLSEKVV LRRKLKSFQH QDMRLKNVLK IKLSGLMDLF KNRYKRFRKP PIDDAFLVHL FEPETSNDPN WQPAYIKDEN MILEVSTGRK FFNMDLDIVE ERLWNGYYSE PKQFLKDIEL IYRDANTIGD RERVIKASEM FANAQMGIEE ISTPDFIQEC KATRQRDLER QELFLEDEEK RAAMELEAKE QSQENILQEP DLKDNKANEF GVAAGNQLQA QLQTTINTAS IVNNSEVPQP IDTNLYKKEI PAAIPSAVDK EKAVIPEDSG ANEEYTTELI QATCTSEITT DDDERARKEP KENEDSLQTQ VTEENFSKID ANTNNINHVK EIQSVNKPNS LHETVEKRER SPIPKEVVEP EQGKKSDKEL ILTPEQIKKV SACLIEHCQN FTVSQLEDVH SSVAKIIWKS KSAWDKTGTV DEIIKFLSE UniProtKB: ATPase histone chaperone YTA7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
Details: Solution was made fresh and detergent was added to solve a preference orientation issue. | ||||||||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Support film - Material: GOLD / Support film - topology: CONTINUOUS | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV / Details: Blot 3 seconds, blot force 3. | ||||||||||||||||||||||||
| Details | The sample was a novel chromatin remodeler and an AAA+ ATPase. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 193.0 K / Max: 193.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.0 e/Å2 Details: A total of 75 frames was recorded for each micrograph stack. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)