[English] 日本語
 Yorodumi
Yorodumi- EMDB-28780: Eilat virus/Eastern equine encephalitis virus chimeric vaccine ca... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Eilat virus/Eastern equine encephalitis virus chimeric vaccine candidate | ||||||||||||||||||
 Map data Map data | T=4 common-lines map | ||||||||||||||||||
 Sample Sample | Eilat virus/Eastern equine encephalitis virus chimeric virus != synthetic viruses Eilat virus/Eastern equine encephalitis virus chimeric virus
| ||||||||||||||||||
 Keywords Keywords | alphavirus / togavirus / Togaviridae / EEEV / encephalitis / virus / virion / arbovirus | ||||||||||||||||||
| Biological species |   Eastern equine encephalitis virus / Eastern equine encephalitis virus /  synthetic viruses synthetic viruses | ||||||||||||||||||
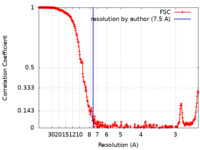
| Method | single particle reconstruction / cryo EM / Resolution: 7.5 Å | ||||||||||||||||||
 Authors Authors | Kaelber JT / Chmielewski D / Chiu W / Auguste AJ | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: Alphavirus Particles Can Assemble with an Alternate Triangulation Number. Authors: Jason T Kaelber / David Chmielewski / Wah Chiu / Albert J Auguste /  Abstract: Alphaviruses are spherical, enveloped RNA viruses primarily transmitted by mosquitoes, and cause significant arthritogenic and neurotropic disease in humans and livestock. Previous reports have shown ...Alphaviruses are spherical, enveloped RNA viruses primarily transmitted by mosquitoes, and cause significant arthritogenic and neurotropic disease in humans and livestock. Previous reports have shown that-in contrast to prototypical icosahedral viruses-alphaviruses incorporate frequent defects, and these may serve important functions in the viral life cycle. We confirm the genus-wide pleomorphism in live viral particles and extend our understanding of alphavirus assembly through the discovery of an alternate architecture of Eastern equine encephalitis virus (EEEV) particles. The alternate = 3 icosahedral architecture differs in triangulation number from the classic = 4 icosahedral organization that typifies alphaviruses, but the alternate architecture maintains the quasi-equivalence relationship of asymmetric units. The fusion spike glycoproteins are more loosely apposed in the = 3 form with corresponding changes in the underlying capsid protein lattice. This alternate architecture could potentially be exploited in engineering alphavirus-based particles for delivery of alphaviral or other RNA. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28780.map.gz emd_28780.map.gz | 671.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28780-v30.xml emd-28780-v30.xml emd-28780.xml emd-28780.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28780_fsc.xml emd_28780_fsc.xml | 25.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28780.png emd_28780.png | 238.1 KB | ||
| Filedesc metadata |  emd-28780.cif.gz emd-28780.cif.gz | 6.6 KB | ||
| Others |  emd_28780_additional_1.map.gz emd_28780_additional_1.map.gz emd_28780_half_map_1.map.gz emd_28780_half_map_1.map.gz emd_28780_half_map_2.map.gz emd_28780_half_map_2.map.gz | 288.4 MB 1.3 GB 1.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28780 http://ftp.pdbj.org/pub/emdb/structures/EMD-28780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28780 | HTTPS FTP |
-Validation report
| Summary document |  emd_28780_validation.pdf.gz emd_28780_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28780_full_validation.pdf.gz emd_28780_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_28780_validation.xml.gz emd_28780_validation.xml.gz | 33.6 KB | Display | |
| Data in CIF |  emd_28780_validation.cif.gz emd_28780_validation.cif.gz | 45.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28780 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28780 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28780 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28780 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28780.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28780.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T=4 common-lines map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.284 Å | ||||||||||||||||||||||||||||||||||||
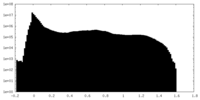
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: T=3 sub-map
| File | emd_28780_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T=3 sub-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
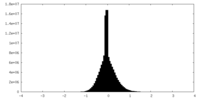
| Density Histograms |
-Half map: odd half-map, unmasked unfiltered
| File | emd_28780_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | odd half-map, unmasked unfiltered | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: even half-map, unmasked unfiltered
| File | emd_28780_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | even half-map, unmasked unfiltered | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Eilat virus/Eastern equine encephalitis virus chimeric virus
| Entire | Name: Eilat virus/Eastern equine encephalitis virus chimeric virus |
|---|---|
| Components |
|
-Supramolecule #1: synthetic viruses
| Supramolecule | Name: synthetic viruses / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Viral genome contains EEEV-FL 93-939 structural proteins and Eilat virus nonstructural proteins. Chimeric virus described in "Novel Insect-Specific Eilat Virus-Based Chimeric Vaccine ...Details: Viral genome contains EEEV-FL 93-939 structural proteins and Eilat virus nonstructural proteins. Chimeric virus described in "Novel Insect-Specific Eilat Virus-Based Chimeric Vaccine Candidates Provide Durable, Mono- and Multivalent, Single-Dose Protection against Lethal Alphavirus Challenge" by Erasmus et al. NCBI-ID: 512285 / Sci species name: synthetic viruses / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / T number (triangulation number): 4 |
-Macromolecule #1: EEEV nucleocapsid protein
| Macromolecule | Name: EEEV nucleocapsid protein / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus / Strain: FL93-939 Eastern equine encephalitis virus / Strain: FL93-939 |
| Sequence | String: CMKLESDKTF PIMLNGQVNG YACVVGGRMF KPLHVEGRID NEQLAAIKLK KASIYDLEYG DVPQCMKSDT LQYTSDKPP GFYNWHHGAV QYENNRFTVP RGVGGKGDSG RPILDNKGRV VAIVLGGVNE GSRTALSVVT W NQKGVTVK DTPEGSEPW GENBANK: GENBANK: AAF04796.1 |
-Macromolecule #2: EEEV glycoprotein E1
| Macromolecule | Name: EEEV glycoprotein E1 / type: protein_or_peptide / ID: 2 / Enantiomer: DEXTRO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus / Strain: FL93-939 Eastern equine encephalitis virus / Strain: FL93-939 |
| Sequence | String: KADDTLQVLN YLWNNNQNFF WMQTLIPLAA LIVSMRMLRC LFCCGPAFLL VCGALGAAAY EHTAVMPNKV GIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCSS KPHPDYQCQV F TGVYPFMW GGAYCFCDTE NTQMSEAYVE ...String: KADDTLQVLN YLWNNNQNFF WMQTLIPLAA LIVSMRMLRC LFCCGPAFLL VCGALGAAAY EHTAVMPNKV GIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCSS KPHPDYQCQV F TGVYPFMW GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NG ETPAKIG DAKLIIGPLS SAWSPFDNKV VVYGHEVYNY DFPEYGTGKA GSFGDLQSRT STSNDLYANT NLK LQRPQA GIVHTPFTQA PSGFERWKRD KGAPLNDVAP FGCSIALEPL RAENCAVGSI PISIDIPDAA FTRI SETPT VSDLECKITE CTYASDFGGI ATVAYKSSKA GNCPIHSPSG VAVIKENDVT LAESGSFTFH FSTAN IHPA FKLQVCTSAV TCKGDCKPPK DHIVDYPAQH TDLLTSAISA TAWSWLKVLV GGTSAFIVLG LIATAV VAL VLF |
-Macromolecule #3: EEEV glycoprotein E2
| Macromolecule | Name: EEEV glycoprotein E2 / type: protein_or_peptide / ID: 3 / Enantiomer: DEXTRO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Sequence | String: PYIADCPNCG HSRCDSPIAI EEVRGDAHAG AIRIQTSAMF GLKTDGVDLA YMSFMNGKTQ KSIKIDNLHV RTSAPCSLV SHHGYYILAQ CPPGDTVTVG FHDGPNRHTC TVAHKVEFRP VGREEYRHPP EHGVELPCNR Y THKRADQG HYVEMHQPGL VADHSLLSIH ...String: PYIADCPNCG HSRCDSPIAI EEVRGDAHAG AIRIQTSAMF GLKTDGVDLA YMSFMNGKTQ KSIKIDNLHV RTSAPCSLV SHHGYYILAQ CPPGDTVTVG FHDGPNRHTC TVAHKVEFRP VGREEYRHPP EHGVELPCNR Y THKRADQG HYVEMHQPGL VADHSLLSIH SAKVKITVPS GAQVKYYCKC PDVREGITSS DHTTTCTDVK QC RAYLIDN KKWVYNFGRL LRGEGDTFKG KLHVPFVPVK AKCIATLAPE LLVEHKHRTL ILHLHPDHPT LLT TRSLGS DANPIRQWIE RPTTVNFTVT GEGLEYTWGN HPPKRVWAQE SGEGNPHGWP HEVVVYYYNR YPLT TIIGL CTCVAIIMVS CVTSVWLLCR ARNLCITPYK LAPNAQVPIL LALLCC |
-Macromolecule #4: EEEV glycoprotein E3
| Macromolecule | Name: EEEV glycoprotein E3 / type: protein_or_peptide / ID: 4 / Enantiomer: DEXTRO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus / Strain: FL93-939 Eastern equine encephalitis virus / Strain: FL93-939 |
| Sequence | String: MCVLANITFP CDQPPCMPCC YEKNPHETLT MLEQNYDSRT YDQLLDAAVK CNARRTRR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: "TEN buffer": 0.05 M Tris-HCl [pH 7.4], 0.1 M NaCl, 0.001 M EDTA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Temperature | Min: 86.8 K |
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 4 / Number real images: 2068 / Average exposure time: 6.0 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.1 mm / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)