+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Huntingtin C-HEAT domain in complex with HAP40 | ||||||||||||
 Map data Map data | Sharpened map used for modelling | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Scaffold protein / HTT / Huntingtin / HAP40 / Huntingtin-associated protein 40 kDa / STRUCTURAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationvesicle cytoskeletal trafficking / : / positive regulation of CAMKK-AMPK signaling cascade / microtubule-based transport / vocal learning / negative regulation of proteasomal protein catabolic process / regulation of CAMKK-AMPK signaling cascade / positive regulation of mitophagy / profilin binding / positive regulation of cilium assembly ...vesicle cytoskeletal trafficking / : / positive regulation of CAMKK-AMPK signaling cascade / microtubule-based transport / vocal learning / negative regulation of proteasomal protein catabolic process / regulation of CAMKK-AMPK signaling cascade / positive regulation of mitophagy / profilin binding / positive regulation of cilium assembly / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / vesicle transport along microtubule / positive regulation of aggrephagy / positive regulation of lipophagy / Golgi organization / dynein intermediate chain binding / phosphoprotein phosphatase activity / establishment of mitotic spindle orientation / dynactin binding / Regulation of MECP2 expression and activity / postsynaptic cytosol / beta-tubulin binding / presynaptic cytosol / heat shock protein binding / inclusion body / centriole / autophagosome / cytoplasmic vesicle membrane / negative regulation of extrinsic apoptotic signaling pathway / protein destabilization / kinase binding / p53 binding / late endosome / transmembrane transporter binding / early endosome / nuclear body / positive regulation of apoptotic process / axon / apoptotic process / dendrite / perinuclear region of cytoplasm / endoplasmic reticulum / Golgi apparatus / protein-containing complex / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
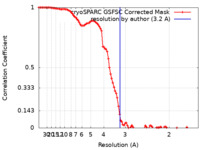
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Harding RJ / Deme JC / Alteen MG / Arrowsmith CH / Lea SM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Expanding the Huntingtons disease research toolbox; validated huntingtin subdomain constructs for biochemical and structural investigation of the huntingtin protein Authors: Alteen MG / Deme JC / Alvarez CP / Loppnau P / Hutchinson A / Seitova A / Chandrasekaran R / Silva Ramos E / Secker C / Alqazzaz M / Wanker EE / Lea SM / Arrowsmith CH / Harding RJ | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28766.map.gz emd_28766.map.gz | 85.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28766-v30.xml emd-28766-v30.xml emd-28766.xml emd-28766.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28766_fsc.xml emd_28766_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_28766.png emd_28766.png | 58.5 KB | ||
| Masks |  emd_28766_msk_1.map emd_28766_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28766.cif.gz emd-28766.cif.gz | 5.9 KB | ||
| Others |  emd_28766_additional_1.map.gz emd_28766_additional_1.map.gz emd_28766_half_map_1.map.gz emd_28766_half_map_1.map.gz emd_28766_half_map_2.map.gz emd_28766_half_map_2.map.gz | 79.9 MB 84.3 MB 84.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28766 http://ftp.pdbj.org/pub/emdb/structures/EMD-28766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28766 | HTTPS FTP |
-Validation report
| Summary document |  emd_28766_validation.pdf.gz emd_28766_validation.pdf.gz | 837.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28766_full_validation.pdf.gz emd_28766_full_validation.pdf.gz | 836.7 KB | Display | |
| Data in XML |  emd_28766_validation.xml.gz emd_28766_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_28766_validation.cif.gz emd_28766_validation.cif.gz | 22.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28766 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28766 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28766 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28766 | HTTPS FTP |
-Related structure data
| Related structure data |  8sahM M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28766.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28766.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map used for modelling | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28766_msk_1.map emd_28766_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
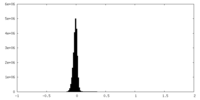
| Density Histograms |
-Additional map: Additional Map
| File | emd_28766_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
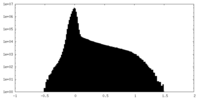
| Density Histograms |
-Half map: Half Map 1
| File | emd_28766_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
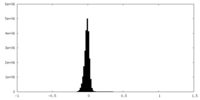
| Density Histograms |
-Half map: Half Map 2
| File | emd_28766_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
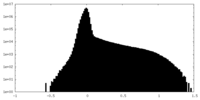
| Density Histograms |
- Sample components
Sample components
-Entire : Full-length Huntingtin-HAP40 complex from subdomain fragments
| Entire | Name: Full-length Huntingtin-HAP40 complex from subdomain fragments |
|---|---|
| Components |
|
-Supramolecule #1: Full-length Huntingtin-HAP40 complex from subdomain fragments
| Supramolecule | Name: Full-length Huntingtin-HAP40 complex from subdomain fragments type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 390 KDa |
-Macromolecule #1: Huntingtin
| Macromolecule | Name: Huntingtin / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVSPDKDWYV HLVKSQCWTR SDSALLEGAE LVNRIPAEDM NAFMMNSEFN LSLLAPCLSL GMSEISGGQK SALFEAAREV TLARVSGTVQ QLPAVHHVFQ PELPAEPAAY WSKLNDLFGD AALYQSLPTL ARALAQYLVV VSKLPSHLHL PPEKEKDIVK FVVATLEALS ...String: MVSPDKDWYV HLVKSQCWTR SDSALLEGAE LVNRIPAEDM NAFMMNSEFN LSLLAPCLSL GMSEISGGQK SALFEAAREV TLARVSGTVQ QLPAVHHVFQ PELPAEPAAY WSKLNDLFGD AALYQSLPTL ARALAQYLVV VSKLPSHLHL PPEKEKDIVK FVVATLEALS WHLIHEQIPL SLDLQAGLDC CCLALQLPGL WSVVSSTEFV THACSLIHCV HFILEAVAVQ PGEQLLSPER RTNTPKAISE EEEEVDPNTQ NPKYITAACE MVAEMVESLQ SVLALGHKRN SGVPAFLTPL LRNIIISLAR LPLVNSYTRV PPLVWKLGWS PKPGGDFGTA FPEIPVEFLQ EKEVFKEFIY RINTLGWTSR TQFEETWATL LGVLVTQPLV MEQEESPPEE DTERTQINVL AVQAITSLVL SAMTVPVAGN PAVSCLEQQP RNKPLKALDT RFGRKLSIIR GIVEQEIQAM VSKRENIATH HLYQAWDPVP SLSPATTGAL ISHEKLLLQI NPERELGSMS YKLGQVSIHS VWLGNSITPL REEEWDEEEE EEADAPAPSS PPTSPVNSRK HRAGVDIHSC SQFLLELYSR WILPSSSARR TPAILISEVV RSLLVVSDLF TERNQFELMY VTLTELRRVH PSEDEILAQY LVPATCKAAA VLGMDKAVAE PVSRLLESTL RSSHLPSRVG ALHGILYVLE CDLLDDTAKQ LIPVISDYLL SNLKGIAHCV NIHSQQHVLV MCATAFYLIE NYPLDVGPEF SASIIQMCGV MLSGSEESTP SIIYHCALRG LERLLLSEQL SRLDAESLVK LSVDRVNVHS PHRAMAALGL MLTCMYTGKE KVSPGRTSDP NPAAPDSESV IVAMERVSVL FDRIRKGFPC EARVVARILP QFLDDFFPPQ DIMNKVIGEF LSNQQPYPQF MATVVYKVFQ TLHSTGQSSM VRDWVMLSLS NFTQRAPVAM ATWSLSCFFV SASTSPWVAA ILPHVISRMG KLEQVDVNLF CLVATDFYRH QIEEELDRRA FQSVLEVVAA PGSPYHRLLT CLRNVGGSGD YKDDDDK |
-Macromolecule #2: 40-kDa huntingtin-associated protein
| Macromolecule | Name: 40-kDa huntingtin-associated protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHHHHHSSG RENLYFQGMA AAAAGLGGGG AGPGPEAGDF LARYRLVSNK LKKRFLRKPN VAEAGEQFGQ LGRELRAQEC LPYAAWCQLA VARCQQALFH GPGEALALTE AARLFLRQER DARQRLVCPA AYGEPLQAAA SALGAAVRLH LELGQPAAAA ALCLELAAAL ...String: MHHHHHHSSG RENLYFQGMA AAAAGLGGGG AGPGPEAGDF LARYRLVSNK LKKRFLRKPN VAEAGEQFGQ LGRELRAQEC LPYAAWCQLA VARCQQALFH GPGEALALTE AARLFLRQER DARQRLVCPA AYGEPLQAAA SALGAAVRLH LELGQPAAAA ALCLELAAAL RDLGQPAAAA GHFQRAAQLQ LPQLPLAALQ ALGEAASCQL LARDYTGALA VFTRMQRLAR EHGSHPVQSL PPPPPPAPQP GPGATPALPA ALLPPNSGSA APSPAALGAF SDVLVRCEVS RVLLLLLLQP PPAKLLPEHA QTLEKYSWEA FDSHGQESSG QLPEELFLLL QSLVMATHEK DTEAIKSLQV EMWPLLTAEQ NHLLHLVLQE TISPSGQGV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 51.3 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.4 µm / Nominal defocus min: 2.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)