[English] 日本語
 Yorodumi
Yorodumi- EMDB-28667: Cryo-EM map of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1-prime | |||||||||
 Map data Map data | Unsharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
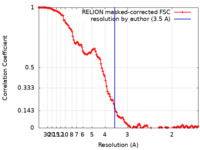
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Li Y / Langley C / Azumaya CM / Echeverria I / Chesarino NM / Emerman M / Cheng Y / Gross JD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: The structural basis for HIV-1 Vif antagonism of human APOBEC3G. Authors: Yen-Li Li / Caroline A Langley / Caleigh M Azumaya / Ignacia Echeverria / Nicholas M Chesarino / Michael Emerman / Yifan Cheng / John D Gross /  Abstract: The APOBEC3 (A3) proteins are host antiviral cellular proteins that hypermutate the viral genome of diverse viral families. In retroviruses, this process requires A3 packaging into viral particles. ...The APOBEC3 (A3) proteins are host antiviral cellular proteins that hypermutate the viral genome of diverse viral families. In retroviruses, this process requires A3 packaging into viral particles. The lentiviruses encode a protein, Vif, that antagonizes A3 family members by targeting them for degradation. Diversification of A3 allows host escape from Vif whereas adaptations in Vif enable cross-species transmission of primate lentiviruses. How this 'molecular arms race' plays out at the structural level is unknown. Here, we report the cryogenic electron microscopy structure of human APOBEC3G (A3G) bound to HIV-1 Vif, and the hijacked cellular proteins that promote ubiquitin-mediated proteolysis. A small surface explains the molecular arms race, including a cross-species transmission event that led to the birth of HIV-1. Unexpectedly, we find that RNA is a molecular glue for the Vif-A3G interaction, enabling Vif to repress A3G by ubiquitin-dependent and -independent mechanisms. Our results suggest a model in which Vif antagonizes A3G by intercepting it in its most dangerous form for the virus-when bound to RNA and on the pathway to packaging-to prevent viral restriction. By engaging essential surfaces required for restriction, Vif exploits a vulnerability in A3G, suggesting a general mechanism by which RNA binding helps to position key residues necessary for viral antagonism of a host antiviral gene. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28667.map.gz emd_28667.map.gz | 131.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28667-v30.xml emd-28667-v30.xml emd-28667.xml emd-28667.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28667_fsc.xml emd_28667_fsc.xml | 12.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_28667.png emd_28667.png | 50.1 KB | ||
| Masks |  emd_28667_msk_1.map emd_28667_msk_1.map | 166.4 MB |  Mask map Mask map | |
| Others |  emd_28667_additional_1.map.gz emd_28667_additional_1.map.gz emd_28667_half_map_1.map.gz emd_28667_half_map_1.map.gz emd_28667_half_map_2.map.gz emd_28667_half_map_2.map.gz | 141.3 MB 131.6 MB 131.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28667 http://ftp.pdbj.org/pub/emdb/structures/EMD-28667 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28667 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28667 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28667.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28667.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
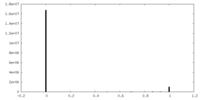
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28667_msk_1.map emd_28667_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
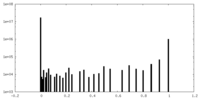
| Density Histograms |
-Additional map: #1
| File | emd_28667_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
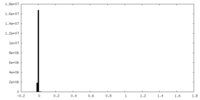
| Density Histograms |
-Half map: #2
| File | emd_28667_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
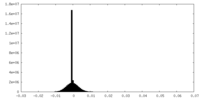
| Density Histograms |
-Half map: #1
| File | emd_28667_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Vif-E3 ligase substrate receptor (VCBC) in complex with hum...
| Entire | Name: HIV-1 Vif-E3 ligase substrate receptor (VCBC) in complex with human APOBEC3G and RNA |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Vif-E3 ligase substrate receptor (VCBC) in complex with hum...
| Supramolecule | Name: HIV-1 Vif-E3 ligase substrate receptor (VCBC) in complex with human APOBEC3G and RNA type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|
-Supramolecule #2: human APOBEC3G
| Supramolecule | Name: human APOBEC3G / type: complex / ID: 2 / Chimera: Yes / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: HIV-1 Vif-E3 ligase substrate receptor (VCBC)
| Supramolecule | Name: HIV-1 Vif-E3 ligase substrate receptor (VCBC) / type: complex / ID: 3 / Chimera: Yes / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.46 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 68.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)